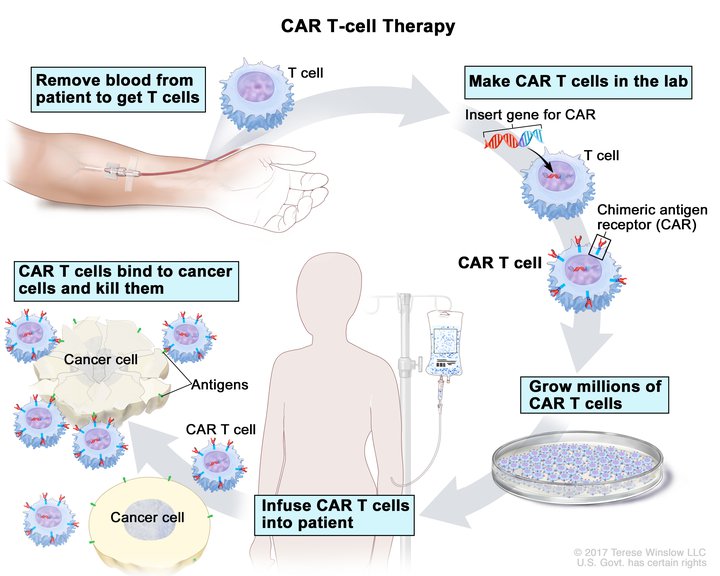
CAR T-cells, which are immune cells reprogrammed to fight cancer, are promising novel cancer therapies to treat certain types of tumors. But, the underlying biological pathways that lead to remission or toxicity are not fully understood.
In a study published in Science Signaling, CPTAC investigator (part of the Proteogenomic Translational Research Centers) Dr. Amanda Paulovich, her team, and fellow researchers at Fred Hutchinson Cancer Research Center used proteomics to profile signaling events in two different, clinically relevant CARs using cell and mouse models.
This paper is the first to use mass spectrometry (MS)-based proteomics to comprehensively analyze proteins involved in CAR T-cell signaling. More importantly, understanding these pathways is crucial for crafting safer, more effective CAR T-cell immunotherapies and reducing toxicity.
The researchers studied the differences between CARs designed with two co-stimulatory domains, CD28 and 4-1BB, which are currently used in clinical trials. Researchers found that similar phosphoprotein signaling events were activated but that the timing and the strength of the signal varied. The CD28 CAR design showed faster, more potent activity while the 4-1BB CAR showed slower, milder, and more persistent activation.
Further testing in a xenograft mouse model revealed that the 4-1BB CAR was more effective in clearing diffuse lymphoma cancer cells compared to the CD28 CAR. As a result, mass spectrometry analysis can provide insights into CAR signaling that can influence future generations of CAR T-cell design and therapies. Based on these initial data, the team is currently building a targeted, multiple reaction monitoring mass spectrometry-based multiplex assay panel that may be a useful tool for optimizing future CAR constructs.

