Immunotherapies are revolutionizing modern cancer care, but at present, many technologies do not accurately quantify the myriad soluble proteins in the tumor microenvironment which impact immunity. This, in turn, contributes to the pervasiveness of patient resistance and immune-related adverse events. In order overcome these hurdles, it is necessary to design more refined protein quantification techniques that complement immunoassays for quantifying human proteins. In their November 2021 report published in Frontiers In Immunology, Whiteaker et al. described the development and characterization of a multiplexed panel (“IO-1 panel”) of immuno-MRM assays designed to quantify immunomodulatory proteins in human tissue biopsies and biofluids.
The assays target 52 peptides (46 proteins) and are part of a larger effort under the Beau Biden National Cancer Moonshot to accelerate scientific discovery in cancer, foster greater collaboration, and improve the sharing of data. These fit-for-purpose assays showed robust analytical performance and the targeted peptides were widely detected in the biospecimens tested (135 tissues and 45 plasma covering 12 tumor types). All resulting characterization data and antibody reagents are publicly available as resources for the research community through the National Cancer Institute’s Clinical Proteomic Tumor Analysis Consortium (CPTAC) Assay Portal and Antibody Portal. Additionally, the monoclonal antibodies generated in this project were tested for use in other applications, such as western blotting and protein array.
In contrast to untargeted mass spectrometry profiling-based proteomics, targeted proteomics focuses the full analytic capacity of the mass spectrometer on pre-selected peptides. In order to perform liquid chromatography-multiple reaction monitoring mass spectrometry (LC-MRM-MS), peptides are released via proteolysis and quantified as stoichiometric surrogates for proteins; an immunoaffinity enrichment step produces immuno-MRM assays that can precisely quantify low abundance proteins and posttranslational modifications. Importantly, by incorporating stable isotope labeled internal standards, MRM assays can be harmonized across laboratories, even at an international scale. In regard to the assay’s significance and utility, Study Leader Dr. Amanda Paulovich writes,
Hundreds of immunomodulatory proteins in the tumor microenvironment sculpt the T cell response to cancer as part of the “cancer-immunity cycle,” and it is critical that we be able to quantify these proteins in clinical and translational research settings to design and deliver improved immunotherapies. This IO-1 assay panel is our first in a series of upcoming multiplex, targeted MRM-MS-based assay panels to quantify immunomodulatory proteins in human biospecimens to support immune-related clinical research, including immuno-oncology, autoimmunity, inflammation, and infectious disease research.
The development of the targeted, multiplexed IO-1 immuno-MRM assay began with the selection of target proteins and phosphorylation sites related to immunomodulatory functions. Target proteins were identified by a panel of experts in immunotherapies. Stoichiometric surrogate peptides for the target proteins were then identified by mining liquid chromatography-tandem mass spectrometry (LC-MS/MS) proteomic and phosphoproteomic datasets for empirical evidence of LC-MS/MS detectability. Peptides were ranked based on the intensity and frequency of empirical observations in the datasets, as well as their chemical and physical properties, and peptides with frequent mutation sites and PTMs were avoided. In total, a list of 52 peptides (including multiple peptides per protein) were selected for assay development, representing 46 proteins and including 5 phosphorylation sites.
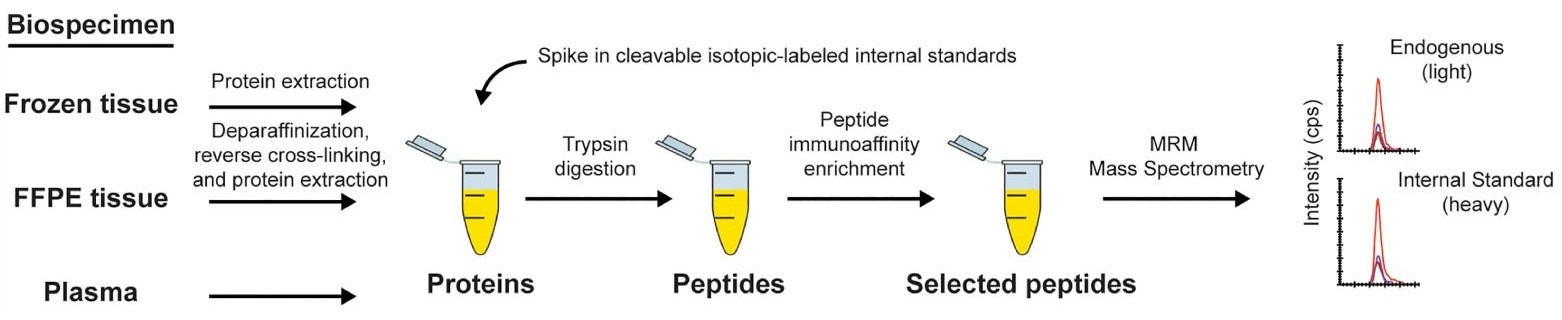
A figure from the publication outlining the immuno-MRM assay workflow, starting from the generation of a protein lysate from the biospecimen of interest and ending when the eluate is analyzed by multiple reaction monitoring-mass spectrometry
The multiplex assay was then applied to a panel of tumor tissue specimens to evaluate the utility of the assay and determine sample requirements for analyte detection in tissue and plasma. The tissue panel included 110 frozen and 25 FFPE biospecimens collected from 12 different tumor types including brain, breast, colorectal, endometrium, head and neck, kidney, lung (squamous cell carcinoma and adenocarcinoma), ovarian, pancreas, and soft tissue sarcoma. Overall, when tested in frozen tissue biospecimens, the assay measured endogenous levels of analytes from 48/52 peptides corresponding to 45 proteins being detected above the lower limit of quantification (LLOQ). Notably, when estimating the minimum amount of tissue needed for the analyte to remain above the LLOQ, the group found the number of peptides detected as the amount of tissue decreases remains within ~80% of total, even with ten-fold less input. This indicates that the assays are amenable to a range of biospecimen sizes and input amounts, which is critical given that clinical biospecimens often have limited available material. In the discussion, Whiteaker et al. also emphasized the significance of this development for use in tandem with existing techniques, like IHC. Immuno-MRM measurements add complementary quantitative information and multiplexability that furthers the potential for useful interpretation of results by pathologists.
The result of this body of work is a powerful tool for protein quantification that is fully accessible to future researchers. On the subject of what their findings contribute to the field of Immunotherapy, Study Leader Dr. Jeffery Whiteaker writes, “Multiplexing the measurement of proteins associated with different aspects of immunity into panels will provide a powerful new tool in the hands of researchers aiming to understand immune cycle mechanisms or develop new treatments for disease.”

