Despite unprecedented advances in the development of targeted- and immuno-therapies for various cancer types, pancreatic ductal adenocarcinoma (PDAC) remains a leading cause of cancer-related death worldwide. A comprehensive characterization of PDAC is critical for the development of new therapies and the improvement of patient outcomes. A key challenge affecting proteogenomic analysis of PDAC is that tumors typically have low neoplastic cellularity and heterogeneous composition. Therefore, an effective technique for the enrichment of neoplastic cells from bulk tumor tissue is crucial for the identification of differentially expressed tumor-associated proteins and genomic alterations.
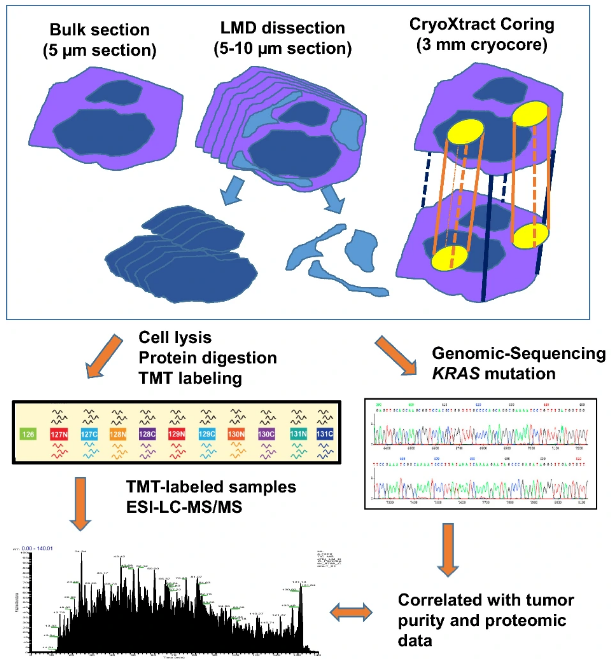
In an effort to overcome this obstacle, a group of researchers from the National Cancer Institute’s Clinical Proteomic Tumor Analysis Consortium (CPTAC), enriched neoplastic cells from bulk tumor using coring and laser microdissection (LMD) sampling techniques; and compared the proteogenomic signatures among samples. The visual abstract from the publication can be seen to the right. Their primary aims, as summarized by Drs. Kay Li and Hui Zhang from the Johns Hopkins University School of Medicine, were to “evaluate the impact of coring and LMD sampling techniques on the observed proteogenomic data, and to address the potential utility of these sampling techniques in the enrichment of neoplastic cells.” This body of work, recently published in Clinical Proteomics, advances the community’s understanding of coring and LMD techniques in the context of integrated proteogenomic characterization of tumor samples.
In the study, the research team used fresh frozen PDAC tumor tissues that contained a variable amount of tumor cellularity and enriched neoplastic cells by coring and LMD sampling techniques. They compared proteogenomic profiles of enriched samples with their corresponding bulk tumor samples without enrichment from the same tumor. These three types of samples were analyzed by TMT-based quantitative proteomics and whole exome genomic sequencing, respectively. The study showed that distinctive proteomic patterns and abundances of certain tumor-associated proteins were revealed in tumor samples obtained by different sampling techniques. Coring sample and bulk tissues had comparable proteome profiles, while LMD samples had the most distinct proteome composition in comparison. Further genomic analysis demonstrated that KRAS mutations were significantly enriched in LMD samples while coring was less effective in this aspect, especially in cases where original bulk tissues contained low neoplastic cellularity. The study offered that, in PDAC, both bulk and coring sampling techniques gave similar proteogenomic signatures, while the greatest enrichment of neoplastic cellularity could be obtained with the LMD technique.