Renal cell carcinoma (RCC) is among the ten most diagnosed cancers worldwide for men and women and comprises a wide array of histologically and genetically defined subtypes involving the kidney. Clear cell RCC (ccRCC) accounts for ~75% of all RCC cases and the majority of renal cancer-associated deaths. High inter- and intra-tumoral heterogeneity in ccRCC represents a key clinical challenge in the molecular characterization of ccRCC and is a confounding factor for therapy selection. In an effort to close this gap, a group of researchers from the National Cancer Institute’s Clinical Proteomic Tumor Analysis Consortium (CPTAC) performed a deep analysis of ccRCC proteogenomic data. Their primary aim, as summarized by Dr. Li Ding from Washington University School of Medicine in St. Louis, was to “identify markers and targets in high grade diseases with aggressive histologic features [in order to] facilitate clinical translation in personalized treatment of ccRCC patients.” Additionally, the researchers rigorously documented phenotypic and genotypic aspects of ccRCC inter- and intra- tumor heterogeneity. This body of research, recently published in Cancer Cell, constitutes a leap forward in our understanding of ccRCC and a landmark achievement in integrated proteogenomic characterization to better predict molecular subtypes.
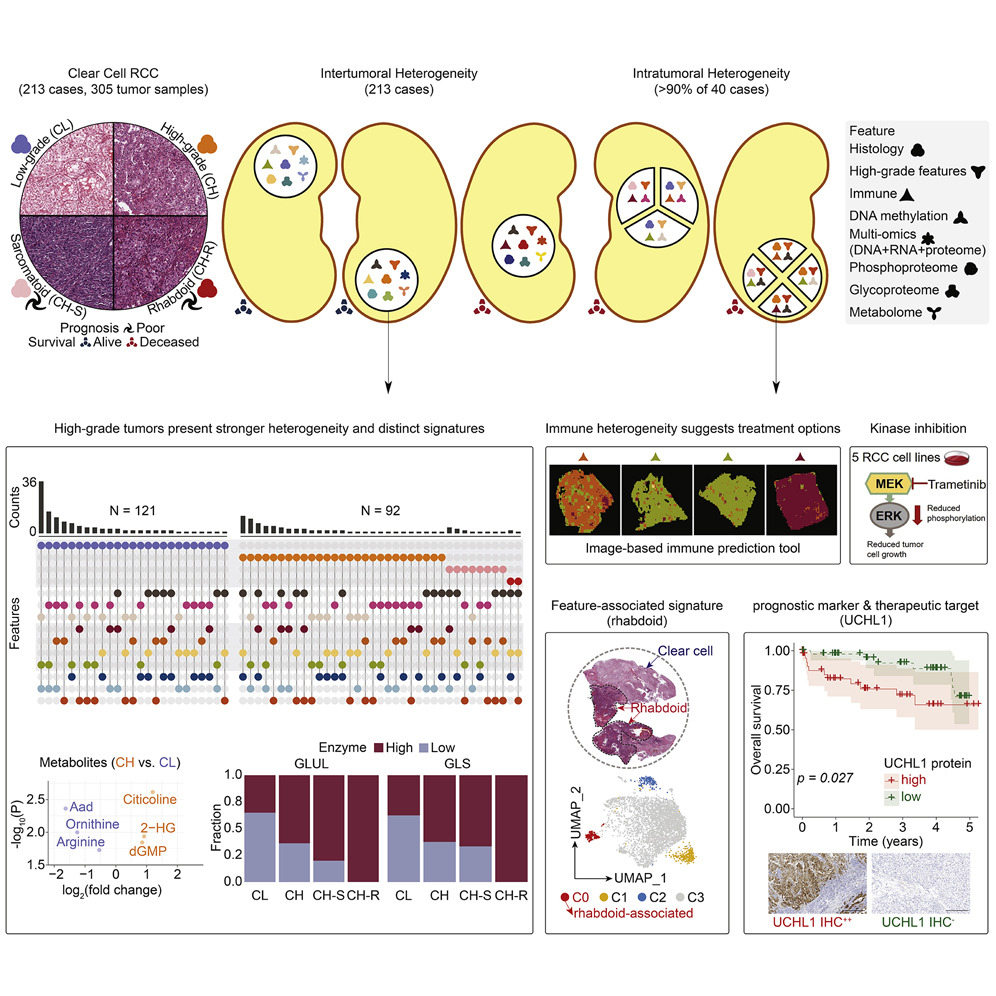
Published by Cell in 2019, the initial CPTAC investigation of ccRCC characterized 103 cases and highlighted a variety of early chromosomal translocation alterations leading to chr3p loss, identified tumor-specific proteomic and phosphoproteomic alterations independent of mRNA expression, and defined specific tumor microenvironment and subtypes based upon mRNAs and proteins. In this new study, the CPTAC ccRCC cohort has been expanded to 305 tumor segments from 213 cases. The subsequent proteogenomic analysis was expanded to include an enhanced histopathologic review of the tumors, proteomic analysis of multi-sampled tumor segments, multi-sample single-nuclei analysis of selected tumors, in vitro analysis of therapeutically targeted tumor-specific phosphoproteomic events, evaluation of tumor-specific metabolites, and a novel analysis of tumor-specific glycoprotein signature. When asked what excites her about the publication, Dr. Hui Zhang from Johns Hopkins University emphasized the proteomic data generated by data-independent generation, all of which is available publicly in the NCI's Proteomic and Genomic Data Commons to aid future biological and clinical investigations.
The extensive multi-omic nature of the study enabled the rigorous validation of high-degree tumor heterogeneity in ccRCC tumors as well as the enrichment of specific proteogenomic features across high-grade tumors. This study identified UCHL1 expression in particular as a candidate biomarker for aggressive ccRCC disease associated with molecular features such as DNA methylation, genome instability, and BAP1 mutation. In conversation with this idea, Dr. Saravana Mohan Dhanasekaran from University of Michigan described this work as similar to a detective in a mystery novel, “using a focused approach to reveal key associations that are hidden in plain sight.” By scouring various ccRCC proteogenomic data mounds generated by the CPTAC consortium, the team was able to identify mRNA/protein culprits associated with different ccRCC histological patterns, discover prognostic/therapeutic markers and study phenotypic and genotypic aspects of ccRCC tumor heterogeneity.
Dr. Dhanasekaran further elaborated “following independent cohort validation, UCHL1 could potentially be developed as a clinical assay to inform future ccRCC treatment strategies. One of our aims is to make a significant impact on decreasing ccRCC mortality by developing a set of clinical tools for the early detection of aggressive cancers.” NIH investigator Dr. Chris Ricketts offered that "this knowledge advances us towards our goal of creating more effect treatment for kidney cancer patients; it benefits and builds upon previous work from many researchers and will hopefully drive further advances in the future.” An increased understanding of accurate and easily evaluable biomarkers for these specific proteogenomic features will be critical for the adaption of targeted combination therapy.

