The following is the first entry in a new Q&A series highlighting selected Clinical Proteomic Tumor Analysis Consortium (CPTAC) researchers and their work. Join us as we discuss molecular and computational techniques in biology with Natalie Clark, PhD from the Proteomics Platform at the Broad Institute. Transcript is edited for clarity.
Q: Can you describe your experience integrating molecular and computational techniques in biology?
NC: I started on this hybrid experimental-computational journey during my PhD at North Carolina State University, where I worked in plant biology. My research utilized a plant called Arabidopsis and during that time I was not only working in the lab … I was also doing data analysis …and I found that I really like to be able to do both. I like to be able to be in the wet lab, do the experiment, take the data, and then do the analysis all by myself--that was really fulfilling for me. I continued that (hybrid role) during my post-doc at Iowa State University (ISU), where I started to do proteomics, and now at the Broad Institute I’ve transitioned into a fully computational role. So, while I don't do any of the experiments anymore, I still have that foundation and I find that it helps me when I do the analysis, or as I develop these analysis methods since I have an idea of the biology and the techniques.
Q: Please elaborate on your role in the CPTAC-LUAD project.
NC: The CPTAC-LUAD (Lung Adenocarcinoma) project at the Broad Institute  involves integrating various genomics and proteomics data sets from different cohorts. This project includes a discovery cohort (Cell, 2020) and a confirmatory cohort… I [joined] after the first cohort had been published (graphical abstract pictured right) and the second cohort was being finalized. My first project was to harmonize these two cohorts because while they were processed and collected in similar ways there was a gap in time… so additional processing was required to ensure they were comparable. Moreover, we're collaborating with the ICPC-Taiwan team and, similarly, [they also have two cohorts]. If you put these four cohorts together, we have over 400 patient samples, or over 800 unique samples when you separate the tumors and the [normal adjacent tissue samples], which is definitely the largest data set that I've ever worked with personally. Integrating everything across different cohorts, across different countries, and across different omes has been challenging but fulfilling and I think at the end of the day, this is going to be a really exciting and intriguing collaboration.
involves integrating various genomics and proteomics data sets from different cohorts. This project includes a discovery cohort (Cell, 2020) and a confirmatory cohort… I [joined] after the first cohort had been published (graphical abstract pictured right) and the second cohort was being finalized. My first project was to harmonize these two cohorts because while they were processed and collected in similar ways there was a gap in time… so additional processing was required to ensure they were comparable. Moreover, we're collaborating with the ICPC-Taiwan team and, similarly, [they also have two cohorts]. If you put these four cohorts together, we have over 400 patient samples, or over 800 unique samples when you separate the tumors and the [normal adjacent tissue samples], which is definitely the largest data set that I've ever worked with personally. Integrating everything across different cohorts, across different countries, and across different omes has been challenging but fulfilling and I think at the end of the day, this is going to be a really exciting and intriguing collaboration.
Q: Can you mention aspects of the development and implementation of any analysis methods you're applying at present?
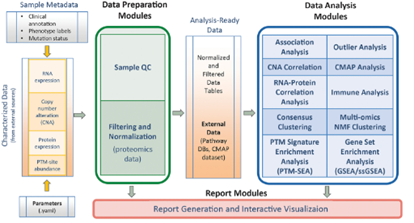
NC: We employ a number of different techniques in our group, and I would say the main goal is to integrate all of these different omics data together. The main technique we use is a suite of tools called PANOPLY developed by Dr. D.R. Mani at the Broad Institute. PANOPLY allows us to normalize, filter, and combine data via methods such as correlation analysis, association analysis, clustering, and outlier analysis (overview pictured below). Additionally, we're working on incorporating new techniques into PANOPLY, such as network inference methods like SC-ION, a method I developed during my post-doc, which infers regulatory networks based on multiple omics types together.
 The development of SC-ION was driven by the need to process large-scale data like you find in transcriptomics and proteomics and use those data to infer unsupervised networks. In essence, you supply [SC-ION] with data, like gene expression or protein expression data, and based on a mathematical model, you can predict if one gene regulates another, potentially with activation or repression. [SC-ION] basically allows you to supply two omes-- one is the regulatory one and the other is the target. It allows you to create networks where you have protein-predicting transcripts or phosphoprotein-predicting proteins, in pretty much any combination you can think of. Additionally, SC-ION provides the network data in a format suitable for visualization in applications like Cytoscape or for further analysis to identify key regulators in the network.
The development of SC-ION was driven by the need to process large-scale data like you find in transcriptomics and proteomics and use those data to infer unsupervised networks. In essence, you supply [SC-ION] with data, like gene expression or protein expression data, and based on a mathematical model, you can predict if one gene regulates another, potentially with activation or repression. [SC-ION] basically allows you to supply two omes-- one is the regulatory one and the other is the target. It allows you to create networks where you have protein-predicting transcripts or phosphoprotein-predicting proteins, in pretty much any combination you can think of. Additionally, SC-ION provides the network data in a format suitable for visualization in applications like Cytoscape or for further analysis to identify key regulators in the network.
Q: Are there any unmet needs or challenges in your field that you hope to resolve during your career or that you foresee for the next generation of young scientists?
NC: One key area is the integration of different omics data. We've made significant progress in combining multiple omics datasets, such as through tools like PANOPLY and SC-ION. However, there are still many methods that claim to be multi-omics that only analyze each omics dataset separately. What we need are more methods that truly integrate multiple omics datasets simultaneously and harmonize them. Another place where I think that we need to innovate is in the single-cell landscape, specifically the single-cell proteomics landscape. It's a challenging area because proteomics data differs in distribution and characteristics compared to genomics data, and there's also a need for optimization on the experimental side.
Q: If a young and aspiring scientist were looking to follow a similar path to yours and reach the point you're at in your career, what advice would you offer them?
NC: My main advice would be to follow the science that interests you. I don't want to advocate and say you need to do the hybrid to do what I do, or that you need to be in the lab and be computational to end up where I am… I think a lot of times people feel pressured that they need to do both. I think that you should do what makes you feel most fulfilled.
That said, no matter where you go in life, I definitely recommend learning a programming language because computer-based analysis is fundamental in today's scientific landscape… and a lot of great resources are available online for free. If possible, I also think a foundation in statistics is really good no matter what you do, whether you're in the lab or doing computational work.
*Transcript edited for clarity*

