Clear cell renal cell carcinoma (ccRCC) is a common type of kidney cancer that originates from the cells of the renal tubules. Recent research has focused on identifying tumor-cell-specific markers to provide mechanistic insights into cancer etiology and support the development of novel targeted therapies, aiming to improve patient outcomes.
In a study recently published in Nature Communications, CPTAC researchers from Washington University in St. Louis combined single-nucleus RNA sequencing (snRNA-seq), single-nucleus ATAC sequencing (snATAC-seq), spatial transcriptomics and matched bulk proteogenomics data to expose the cellular architecture and molecular characteristics of ccRCC. Due to its potential translational impact, the publication was selected to be featured on the Nature Communications Editors’ Highlights webpage.
Through the analysis of snRNA-seq, snATAC-seq, bulk RNA-seq, and proteomics data, researchers identified 324 ccRCC-specific markers. The team prioritized a list of 20 markers based on significantly higher expression in tumor cells compared to normal proximal tubule cells. All but one of these markers showed higher chromatin accessibility in their regulatory regions. Notably, ceruloplasmin (CP), a previously reported marker, was repeatedly rediscovered and validated by data from diverse platforms.
High expression levels of CP in tumor cells were associated with higher tumor grades, shorter overall survival, and a significant difference in CP level between high-grade and low-grade tumors in both the study cohort and the larger ccRCC CPTAC cohort. To investigate CP’s role further, the team generated cell lines with reduced CP expression. Resultant down-regulation of matrisome-associated and inflammatory response genes suggested that CP may mediate tumor-stroma interactions. Based on spatial transcriptomic data, the team concluded that CP may modulate Oncostatin M – oncostatin M receptor signaling between tumor cells and macrophages, as well as other signaling pathways such as TGM2-FN1, associated with cell migration and invasion in cancer. These findings suggest that CP, along with other markers found in the study, could be valuable diagnostic and prognostic tools, offering insights into the etiology of ccRCC and the development of novel targeted therapies.
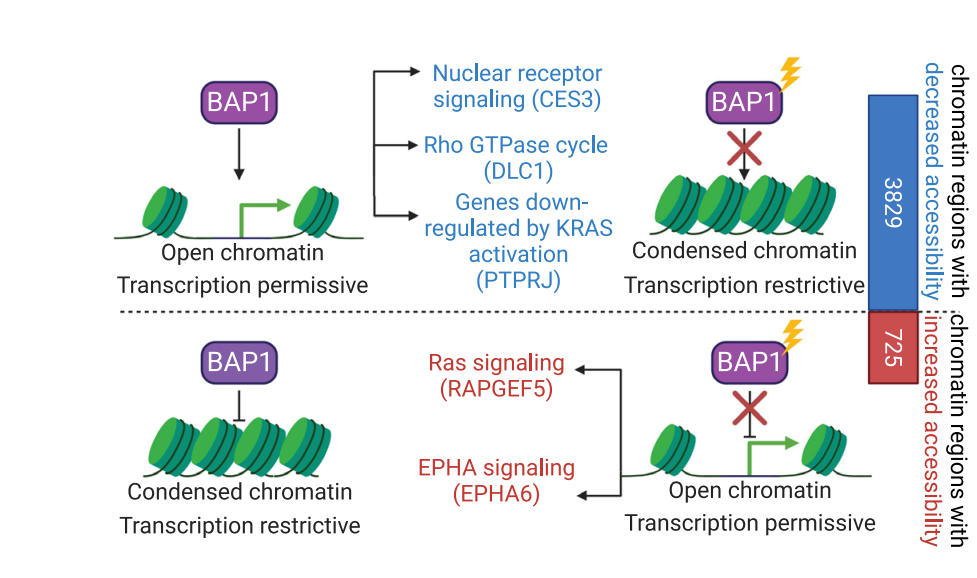
To investigate changes in BAP1 and PBRM1 mutant tumors, with a focus on chromatin accessibility, the team analyzed differentially accessible chromatin regions using snATAC-seq data. They observed that BAP1 deficiency induced chromatin condensation and resulted in more global changes in chromatin accessibility compared to PBRM1 deficiency. In tumors where both were present, BAP1 mutation appeared to have a dominant effect, with those tumors sharing relatively few traits with PBRM1 mutant-only tumors. (Pictured below: schematic diagram showing the differential effects of BAP1 mutations on chromatin accessibility).
The analysis identified differentially expressed genes associated with BAP1 mutations that were most enriched in the nuclear receptor meta-pathway, the RhoAGTPase cycle, and genes downregulated by KRAS activation, among others. Using bulk gene expression and protein datasets, they identified a number of BAP1-specific differentially expressed genes, with CES3 being of particular interest. “CES3 encodes a carboxylesterase involved in lipid metabolism.” Dr. Ding, one of the two senior authors of this paper, said “CES3 down-regulation may promote tumor progression in BAP1-mutants.”
Historically, ccRCC has been recognized for its substantial genetic heterogeneity and abundant genetic alterations. However, whether there are correspondingly high levels of heterogeneity in the epigenome and transcriptome remains largely unknown. This publication from the team at Washington University in St. Louis represents a leap forward in the understanding of ccRCC biology and provides a foundation for further studies aimed at understanding the mechanisms underlying ccRCC tumorigenesis. Dr. Feng Chen, the other senior author of the study concluded, “In addition to contributing to our knowledge of the molecular features and the oncogenic processes of ccRCC, we hope the discoveries described in this paper and the large volume of data generated will empower a wide range of follow-up studies in the RCC community and beyond.”
To learn more, read the full publication at Nature Communications.