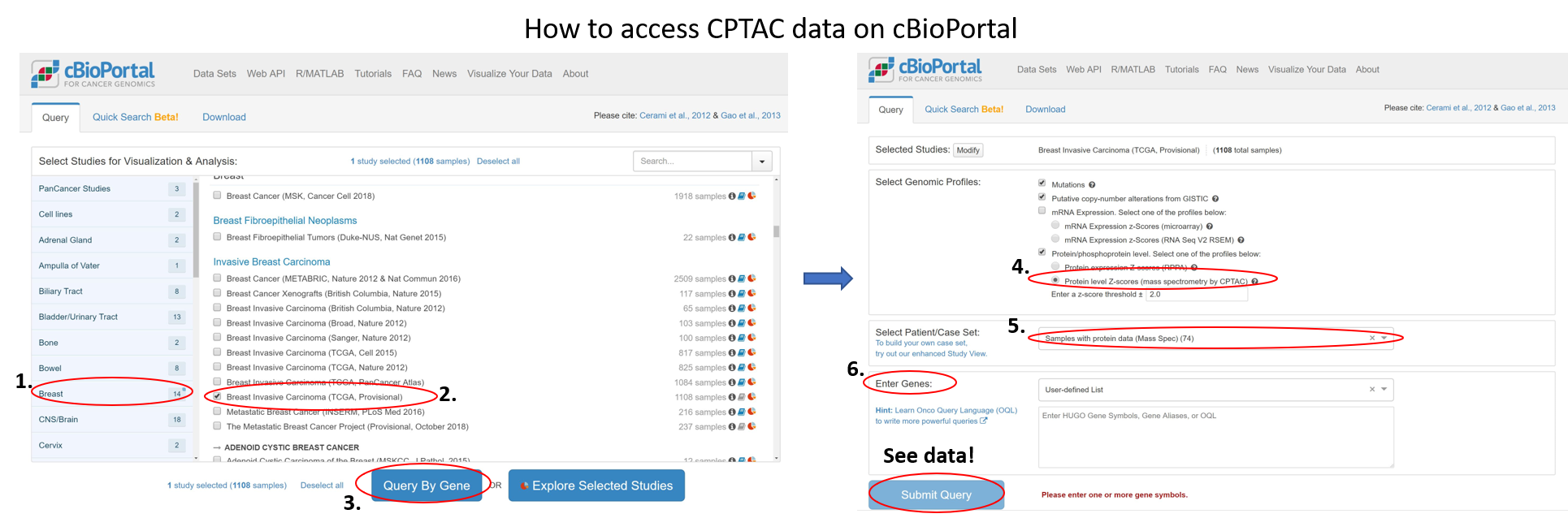
Genomics platform cBioPortal offers a “biologist-friendly” resource for querying mutations, copy number variations and gene expression. It contains data from The Cancer Genome Atlas (TCGA) which includes proteomics data in the form of arrays that use antibodies to track protein levels. CPTAC mass spectrometry-based proteomics, however, has more information than the arrays provide. Researchers at the New York University School of Medicine have integrated CPTAC data from breast, ovarian and colorectal cancer studies.
Integration was performed in several steps. First, data was processed and peptides resulting from mass spectra were identified and categorized. Data from both proteomics and phosphoproteomics was integrated into cBioPortal. Now users can interactively navigate genomics, proteomics and phosphoproteomics together on the platform as explained in the image. Next, researchers added heatmap visualization to a cBioPortal query called OncoPrint. Together, users can search for specific patient samples and obtain gene expression along with protein level expression data.
Other features of integration include enhanced correlation analysis and examining the full set of CPTAC data through computer language R. Overall, CPTAC researchers have taken proteomics data from the breast, ovarian and colorectal studies and transformed it into the cBioPortal platform to give users a broader picture of changes occurring in patient tumors.
This paper was published in Molecular and Cellular Proteomics.