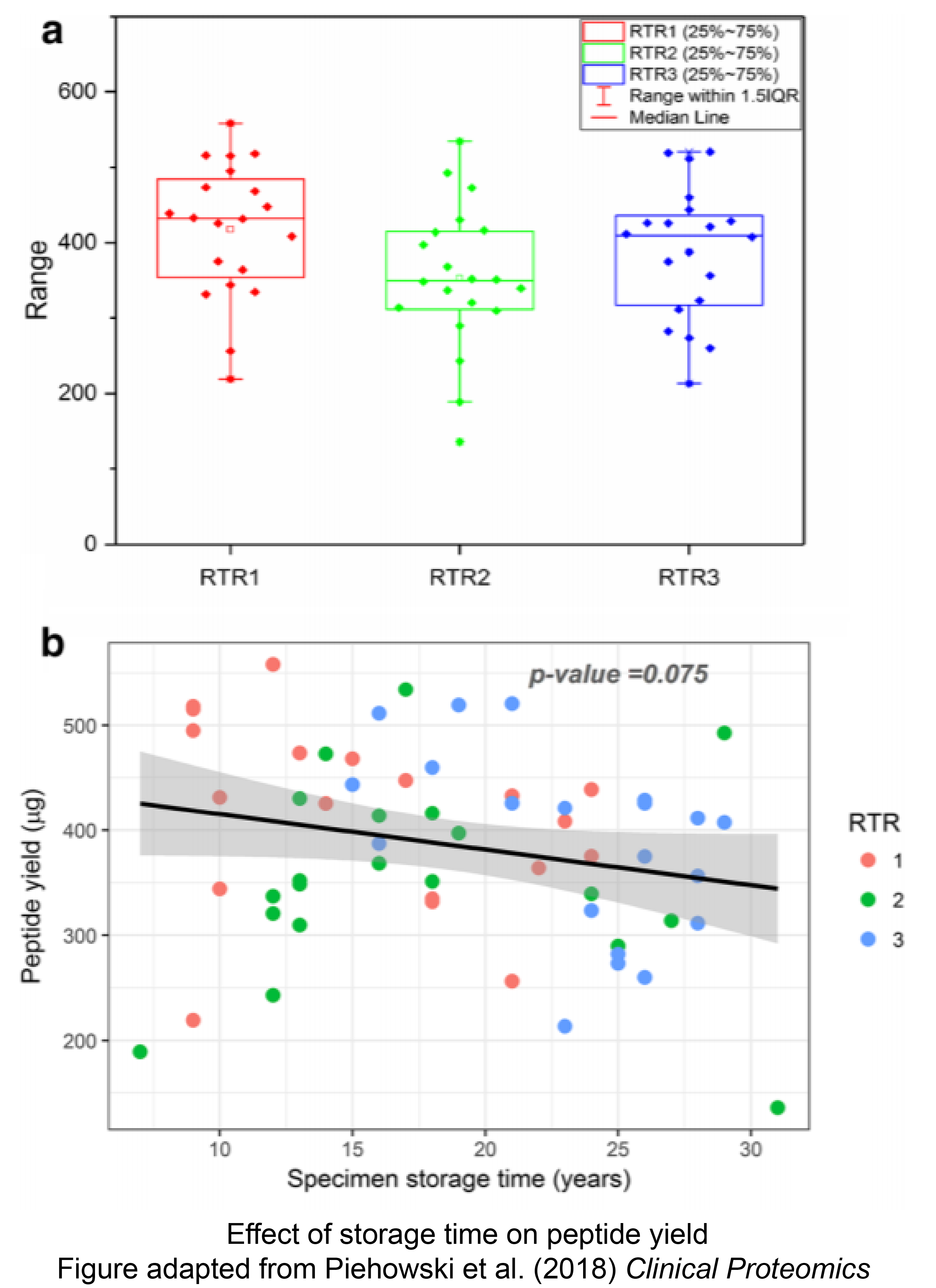
Next-generation mass spectrometry (NGMS) has become a powerful tool for protein identification and quantification from prospectively collected fresh frozen or optimal cutting temperature embedded specimens. However, limitations due to supply, accessibility, and delay of clinical information and outcomes from prospectively collected specimens allow researchers to consider the use of banked specimens. Most biospecimens from NCI’s Surveillance, Epidemiology and End Results (SEER) residual tissue repositories (RTR) are archived and annotated formalin-fixed paraffin-embedded (FFPE) tissue blocks with relevant outcomes information. Due to availability, the reuse of FFPE specimens for deep proteomic characterization with NGMS would be resourceful for population-based cancer studies. The National Cancer Institute’s (NCI’s) Clinical Proteomic Tumor Analysis Consortium (CPTAC), Division of Cancer Treatment and Diagnosis (DCTD) and Division of Cancer Control and Population Science (DCCPS) examined the suitability of the SEER repository tissues for proteomic and phosphoproteimc analysis.
As reported in the journal Clinical Proteomics, the investigators processed 60 FFPE specimens stored from 7 to 32 years for global proteomics and 18 specimens for phosphoproteomics using tandem mass tag (TMT) 10-plex labeling. Additionally, linear modeling and gene set enrichment analysis (GSEA) was used to determine the impact of collection site and storage time. Investigators found that all samples yielded protein mass after extraction for expression analysis. The 18 specimens yielded sufficient mass for phosphopeptide analysis. Although peptide, protein, and phosphopeptide identifications were reduced in FFPE specimens when compared to OCT specimens, there were no statistical differences in protein quantification correlating with collection site or specimen age. Differences between FFPE and OCT embedded specimens were found through GSEA analysis of GO-term level measurements of protein abundance, suggesting that formalin fixation may alter representation of protein categories. This study demonstrates that archived FFPE tissue specimens, with varying storage time and collection sites, serve as a promising source of protein for initial proteomic investigations when paired with rigorous and reproducible NGMS workflows, reducing the need for prospectively collected specimens.

