 It is no coincidence that one cellular process is mentioned time and time again in discussions of cell-signaling pathways in cancer - that is phosphorylation. Since its discovery, it has come to be recognized as a global regulator of many intracellular processes such as growth, proliferation, and cell division. Furthermore, several important aspects of the drug discovery process, including target identification, mechanism of action determination and biomarker identification as well as drug repositioning, require complete understanding of the effects of drugs on protein phosphorylation in relevant biological systems. Thus, any perturbations in the phosphorylation process are likely to drive many of the hallmarks of cancer, such as unchecked cell growth and proliferation.
It is no coincidence that one cellular process is mentioned time and time again in discussions of cell-signaling pathways in cancer - that is phosphorylation. Since its discovery, it has come to be recognized as a global regulator of many intracellular processes such as growth, proliferation, and cell division. Furthermore, several important aspects of the drug discovery process, including target identification, mechanism of action determination and biomarker identification as well as drug repositioning, require complete understanding of the effects of drugs on protein phosphorylation in relevant biological systems. Thus, any perturbations in the phosphorylation process are likely to drive many of the hallmarks of cancer, such as unchecked cell growth and proliferation.
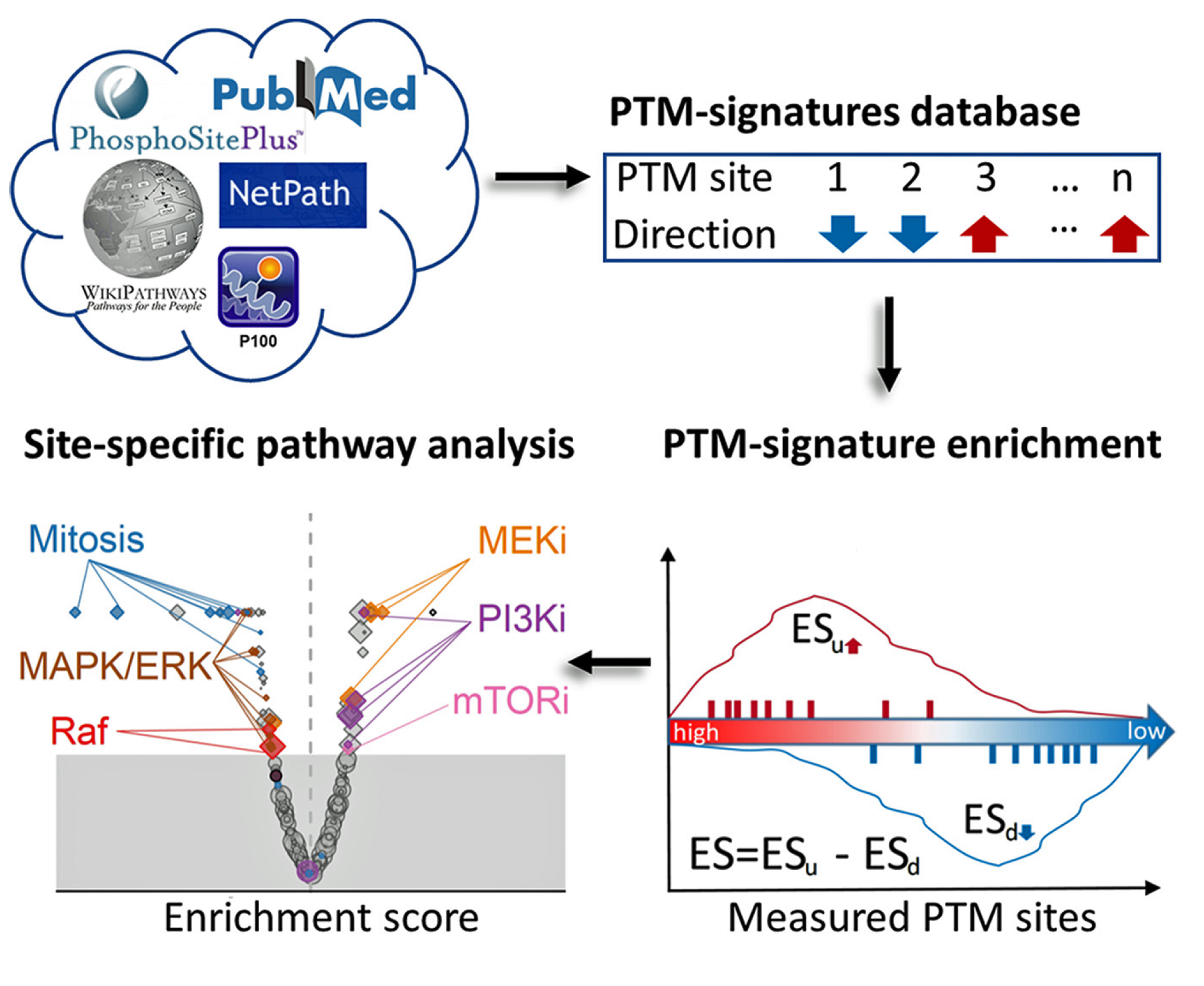
To accelerate research in this area of cancer biology, CPTAC investigators at the Broad Institute along with researchers at UC San Diego, Novartis Institutes for BioMedical Research, Cell Signaling Technology, and others institutions, developed a freely available PTM signatures database (PTMsigDB). PTMsigDB consists of modification site specific signatures of perturbations, kinase activities and signaling pathways curated from more than 2,500 publications, which provides the foundation to perform PTM signature enrichment analysis. This curated resource was then used to perform phosphosite-specific signature analysis for MS-based phosphoproteomics datasets derived from perturbagen studies involving EGF, cell cycle inhibitors and a specific PI3K inhibitor.
A unique feature of PTMsigDB is the annotation of each PTM site with the direction of regulation, i.e. increased or decreased abundance upon a specific perturbation or in the context of signaling through a canonical pathway.
While PTMsigDB enables kinase signature analysis similar to published tools, it additionally includes many curated signature sets of PTM sites mined from PhosphoSitePlus, Netpath, and WikiPathways.
Ultimately the aim is to further extend PTMsigDB when more curated, site-specific molecular pathways are made available by other repositories. Moreover, it is envisioned that this curation process will become a community effort in which researchers studying PTMs in particular pathways will contribute to curating these pathways at the level of PTM sites.
This study was published in Molecular and Cellular Proteomics.

