In a study published on April 21, in Cell Reports Medicine, researchers with the National Cancer Institutes’ (NCI) Clinical Proteomics Tumor Analysis Consortium (CPTAC) take a comprehensive look at the root causes of ovarian high-grade serous carcinoma (HGSC). Through integrated proteomic and genomic analysis of these tumors and comparison to normal tissues, researchers propose new insights about more effective patient drug targets and therapies and reveal factors correlated with patient survival.
HGSC is the most prevalent subtype of ovarian cancer. 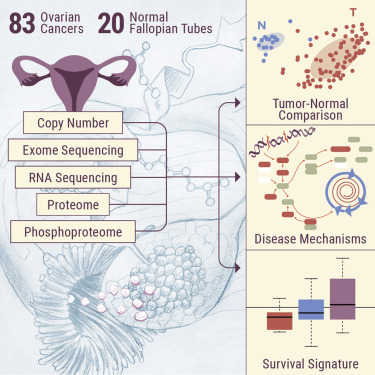 Due to its aggressive behavior and late-stage diagnosis (stages III or IV), it also features the lowest survival rates – only a 29% 5-year survival rate in 2018. Additionally, late-stage HGSC ovarian cancer has a 70-95% chance of recurrence, making it the leading cause of gynecological cancer-related deaths in the developed world.
Due to its aggressive behavior and late-stage diagnosis (stages III or IV), it also features the lowest survival rates – only a 29% 5-year survival rate in 2018. Additionally, late-stage HGSC ovarian cancer has a 70-95% chance of recurrence, making it the leading cause of gynecological cancer-related deaths in the developed world.
This poor prognosis is due to tumor cells with a genome that is very unstable and a genome repair system that is dysfunctional, earning HGSC the title ‘the disease of broken chromosomes’. This combination of high genomic instability and poor repair mechanisms throw the tumor cells into an uncontrolled, rapid growth pattern, with very few effective targets for treatment. Because of this, HGSC standard of care is aggressive, including surgery and chemotherapy with platinum-based drugs that frequently have severe side-effects.
Corresponding authors Karin Rodland and Tao Liu from the Pacific Northwest National Laboratory (PNNL) lead this study, with contributions from CPTAC researchers at John Hopkins Medical Institute, Baylor College of Medicine and Washington University in St. Louis.
Prior to this report, CPTAC researchers and The Cancer Genome Atlas (TCGA) had jointly explored ovarian HGSC in a 2016 proteomic study. Today, the expansion of that study with the inclusion of normal control tissue and robust first-year clinical follow-up data, has revealed even more important features of HGSC ovarian cancer. Using proteogenomic and phosphoproteomic strategies, researchers characterized 83 ovarian HGSC tumor tissues and 20 normal precursor tissues (fallopian tubes) purposefully collected to minimize ischemia time (a necessary step to avoid artifacts in protein phosphorylation related to activation of stress kinases). This led researchers to probe deep into the biology of HGSC to identify several confounding causes of HGSC carcinogenesis. “The design of this CPTAC study allowed us to compare ovarian tumors with the normal tissue that serves as the origin of the cancer – the Fallopian tube epithelium. Comparing normal tissue and tumor tissue allows us to identify what is ‘broken’ in the tumor, and that is an important part of finding a cure. To our knowledge, this is the first comprehensive comparison of Fallopian tube tissues and ovarian tumors at the protein level.” says Dr. Karin Rodland, chief scientist at PNNL.
Identifying the roots of the tumor’s aggressive nature
Using an emerging technique called proteogenomics, researchers studied the central operating mechanism of a tumor – simultaneously looking at changes in its genes, transcripts and proteins. Each layer of this mechanism has an impact on the function of the overall tumor. However, some changes make major impacts, while others make minor ones.
The first prevailing impact pointed to ‘proliferation-induced replication stress’ as a major cause for the severe HGSC genome instability. In short, processes responsible for tumor growth are stuck in the “on” position, causing the cells to continually multiply at an increased rate, and prompting a cellular stress response. As with anything, this high turnover reduces the quality of the product – in this case the genome.
This replication stress was associated with an increase in the signals that turn on the proliferation process – called kinases – and were identified as key drivers of the disease. Several kinases that were specifically implicated include those in the CDK/RB and AURKA proliferation-associated pathways.
The bad news is the present standard of care does not target these pathways. The good news is that these pathways have existing FDA-approved drug therapeutics currently available.
The tumor creates the perfect storm
With one part of the story revealed, scientists turned their attention to the tumors dysfunctional genome repair system – the second arm of its aggression mechanism. A subset of ovarian tumors are deficient in a genome repair mechanism called “homologous recombination”. This deficiency makes tumors sensitive to a particular kind of targeted therapy. CPTAC researchers found a common feature, reduced histone H4 acetylation, among tumors with homologous recombination repair deficiency and proposed that this marker could be exploited to guide therapeutic choices.
Identifying survival in ovarian HGSC patients
Another key outcome from the study was that researchers were able to identify survival-associated signatures between short and long term HGSC survivors. In the 2016 report, scientists derived a panel of 90-proteins whose abundance were highly associated with patient survival. In the current study, tumors with low protein abundance scores strongly correlated with deceased patients and shorter survival times, while patients with longer survival times had an intermediate protein abundance signature. This new knowledge can act as an invaluable resource for clinicians at the time of tumor resectioning to direct the use of targeted therapies and patient management.
According to Dr. Henry Rodriguez, Director of NCI’s Office of Cancer Clinical Proteomic Research (OCCPR), “this study provides new information into the carcinogenesis and progression of HGSC ovarian cancer. The use of proteogenomics to reveal these findings is a testament to the need for a comprehensive approach to cancer research.” OCCPR, through the CPTAC program, has an 8-year history in promoting the development of proteogenomic methods as a reliable tools in the fight against cancer and precision oncology.
This work was supported by grants U24CA160019, U24CA160036 and U24CA210955 from the National Cancer Institute’s (NCI) Clinical Proteomic Tumor Analysis Consortium (CPTAC) and by grant P41GM103493 from the National Institutes of Health. The measurements of proteins and protein activity were done at EMSL, the Environmental Molecular Sciences Laboratory, a user facility funded by the DOE’s Office of Science and located at Pacific Northwest National Laboratory (PNNL).

