According to U.S. National Cancer Institute estimates, approximately 10–20% of all cancer patients will receive a platinum drug over the course of their treatment. Cisplatin, the first platinum-based anti-cancer therapy, was approved by the Food and Drug Administration (FDA) for treating testicular cancer in 1978; subsequently, more platinum formulations (i.e. carboplatin, oxaliplatin) were designed and approved to reduce side effects and bolster results. Unfortunately, in many cases, tumors manage to subvert the cytotoxic effects of platinum, resulting in drug resistance and cancer mortality. In their paper recently published in Oncogene, Huang et al. reviewed over 30 years of research and existing scientific evidence supporting the association of many genes with platinum resistance. This effort has resulted in a manually annotated database of relevant genes for use in large-scale bioinformatic analyses to facilitate the study of platinum resistance and the development of therapies to overcome it.
Understanding and Overcoming Platinum Resistance
The main cytotoxic mechanism of platinum is the generation of mono adducts and crosslinks in DNA. DNA damage caused by platinum results in several cellular responses which, in turn, account for its anti-cancer effect (i.e. severe cellular stress, apoptosis, immune response). Individual tumor responses to platinum treatment are complex, involving both tumor-intrinsic and tumor-extrinsic activities; and the same can be said for the development of resistance. For example, the platinum-resistant PEO4 ovarian cancer cell line is understood to use at least six different strategies to ensure survival,
a) restoration of DNA damage response via a BRCA2 reversion mutation; b) alterations in histone methylation via overexpression of Histone-lysine N-methyltransferase EZH2; c) activation of HIF1A; d) activation of TGFb signaling; e) inhibition of pro-apoptotic Bcl2 family proteins; and f) activation of transcription factor STAT1 by deacetylase HDAC4.
This example represents just one of many challenges facing researchers studying platinum resistance and further demonstrates the value of an annotated database for these genes. At present, bioinformatics algorithms are used to interpret large datasets like the one featured here to provide insights and potentially reveal clinically relevant correlations. Huang et al. cite rapidly expanding bioinformatics datasets created by next generation technologies as a promising tool for understanding complex resistance. As they are developed, new bioinformatic algorithms using a combination of systems biology and artificial intelligence will be able to interpret larger and larger datasets more quickly and accurately.
A Novel Database and Community Resource Describing Platinum Resistance
The gene table featured in the publication was generated via an extensive literature search. The database includes the following information from their review of existing literature: gene and protein identifiers including the NCBI Gene ID, HUGO gene symbol, UniProt protein ID, and other IDs; manually curated, functional annotations related to platinum resistance based on the literature review, with hyperlinked citations; and manually curated annotations indicating whether gene expression is increased or decreased during development of platinum resistance.
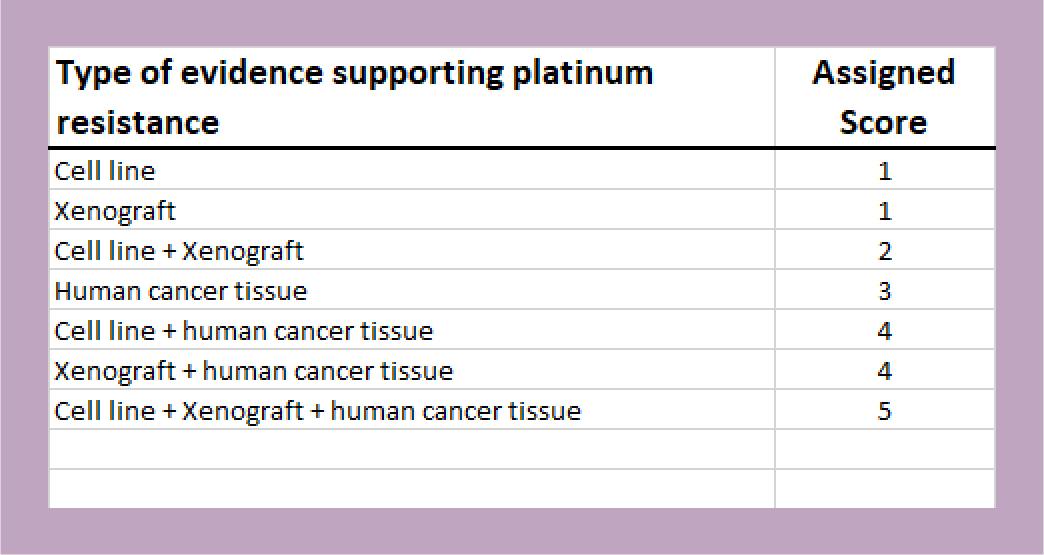 Additionally, the group developed a score to describe the strength of evidence supporting each gene’s role in platinum resistance (see image). This score may be used as a threshold to filter genes based on the level of evidence (cell line, animal model, and/or human tissue).
Additionally, the group developed a score to describe the strength of evidence supporting each gene’s role in platinum resistance (see image). This score may be used as a threshold to filter genes based on the level of evidence (cell line, animal model, and/or human tissue).
The Collaborative Effort and Resulting Significance
This publication represents a collaborative effort between multiple research institutions, including Fred Hutchinson Cancer Research Center, Baylor College of Medicine, Icahn School of Medicine at Mount Sinai, Masonic Cancer Center at the University of Minnesota, and University of Arkansas for Medical Sciences. Study leader Dr. Amanda Paulovich from Fred Hutchinson Cancer Research Center shared her thoughts on the challenges facing platinum response researchers and the importance of this work,
“Despite over 30 years of fantastic literature studying platinum responses in cancer cells, no biomarker to predict platinum response has been clinically translated… The mechanistic complexity of the platinum phenotype likely explains the lack of clinical translation to date, since most predictive biomarker validation studies test one protein at a time by immunohistochemistry, whereas a panel of proteins will likely be required to predict a significant number of resistant cases.”
Dr. Paulovich went on to say the group hopes that the now-public database featured in this study can help develop future research and finally to clinically translate a much-needed multianalyte diagnostic panel to successfully predict platinum responsive tumors. In conversation with Dr. Paulovich, and when asked what makes this work so important, co-author Dr. Tim Star from University of Minnesota said, “Our hope is that this database can be mined by researchers, using sophisticated bioinformatic approaches that include machine learning and artificial intelligence, to better understand the complex response to platinum therapy.” This aspiration is clearly seen in the group’s results, as the current database is fully accessible to the public and its intuitive layout and navigation allow for easy access to information on over 900 genes. The group feels optimistic that proteogenomic investigations of human tumors, along with technologies enabling the measurement of multi-analyte panels, will lead to novel diagnostics for detecting and novel therapies for overcoming platinum resistance.
To view the database, click here: http://ptrc-ddr.cptac-data-view.org
To view the publication in Oncogene, click here: https://www.nature.com/articles/s41388-021-02055-2

