In biomedical research, sample mislabeling or incorrect annotation 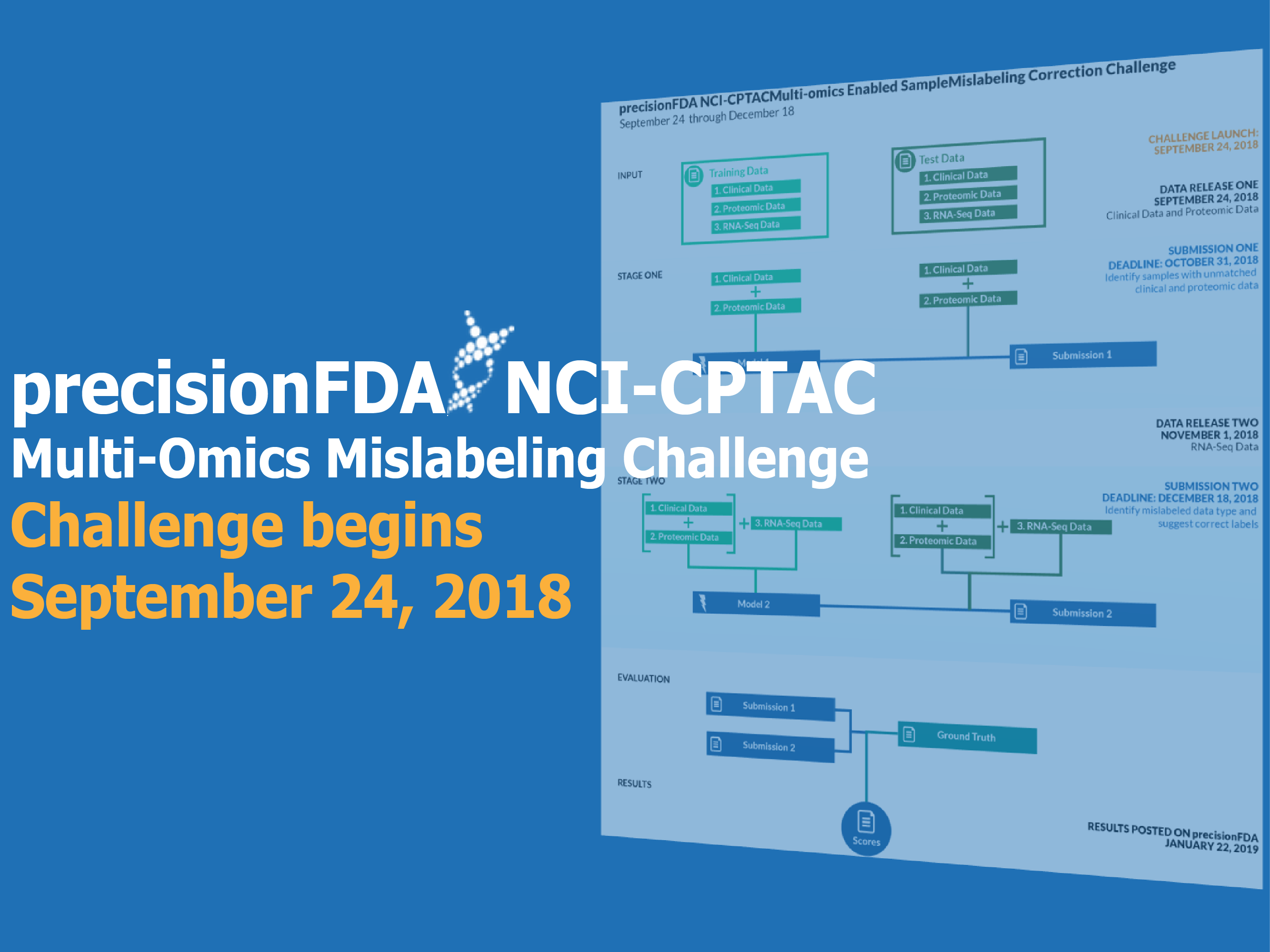 has been a long-standing problem that contributes to irreproducible results and invalid conclusions. These problems are particularly prevalent in large scale multi-omics studies where human errors could arise during sample transferring, sample tracking, large-scale data generation, and data sharing/management. However, the tremendous value of multiple omics studies, as demonstrated in The Cancer Genome Atlas (TCGA) and the Clinical Proteomic Tumor Analysis Consortium (CPTAC) projects, is becoming a powerful approach to precision medicine. Thus, there is a pressing need to develop computational algorithms/tools that can automatically screen for and correct incorrect sample labels or annotations for such studies. This is possible because parallel datasets on the same samples can provide better information to enable identification and correction of mislabeled or misannotated samples than traditional single-omics studies. This crowdsourced precisionFDA NCI-CPTAC challenge is designed to incentivize the development of computational approaches to accurately detect and correct mislabeled samples in large-scale multi-omics studies in two phases.
has been a long-standing problem that contributes to irreproducible results and invalid conclusions. These problems are particularly prevalent in large scale multi-omics studies where human errors could arise during sample transferring, sample tracking, large-scale data generation, and data sharing/management. However, the tremendous value of multiple omics studies, as demonstrated in The Cancer Genome Atlas (TCGA) and the Clinical Proteomic Tumor Analysis Consortium (CPTAC) projects, is becoming a powerful approach to precision medicine. Thus, there is a pressing need to develop computational algorithms/tools that can automatically screen for and correct incorrect sample labels or annotations for such studies. This is possible because parallel datasets on the same samples can provide better information to enable identification and correction of mislabeled or misannotated samples than traditional single-omics studies. This crowdsourced precisionFDA NCI-CPTAC challenge is designed to incentivize the development of computational approaches to accurately detect and correct mislabeled samples in large-scale multi-omics studies in two phases.
Key Dates:
Launch and data release for subchallenge 1: September 24, 2018
Subchallenge 1 submission deadline: October 31, 2018
Data release for subchallenge 2: November 1, 2018
Subchallenge 2 submission deadline: December 18, 2018
Results posted on precisionFDA: January 22, 2019

