Earlier this year, a team from the European Cancer Moonshot Lund Center at Lund University published proteomics and corresponding proteogenomic integration studies for malignant melanomas (MM). The twin publications represent an effort to map protein levels in over 500 MM tumor samples (the MM500 study). By applying deep-learning based single-cell segmentation and phenotyping as well as proteogenomic characterization, the researchers successfully developed a novel Digital Melanoma Pathology. The MM500 study resulted in the identification of more than 15,500 proteoforms via mass spectrometry, from which chromosomal and subcellular localization were annotated within both primary and metastatic melanoma. The consequent molecular map was further bolstered through analysis of blood protein expression, including data on proteins regulated after immunotherapy. In total, this groundbreaking melanoma tumor protein blueprint covers 72% of proteins currently identified in the human proteome.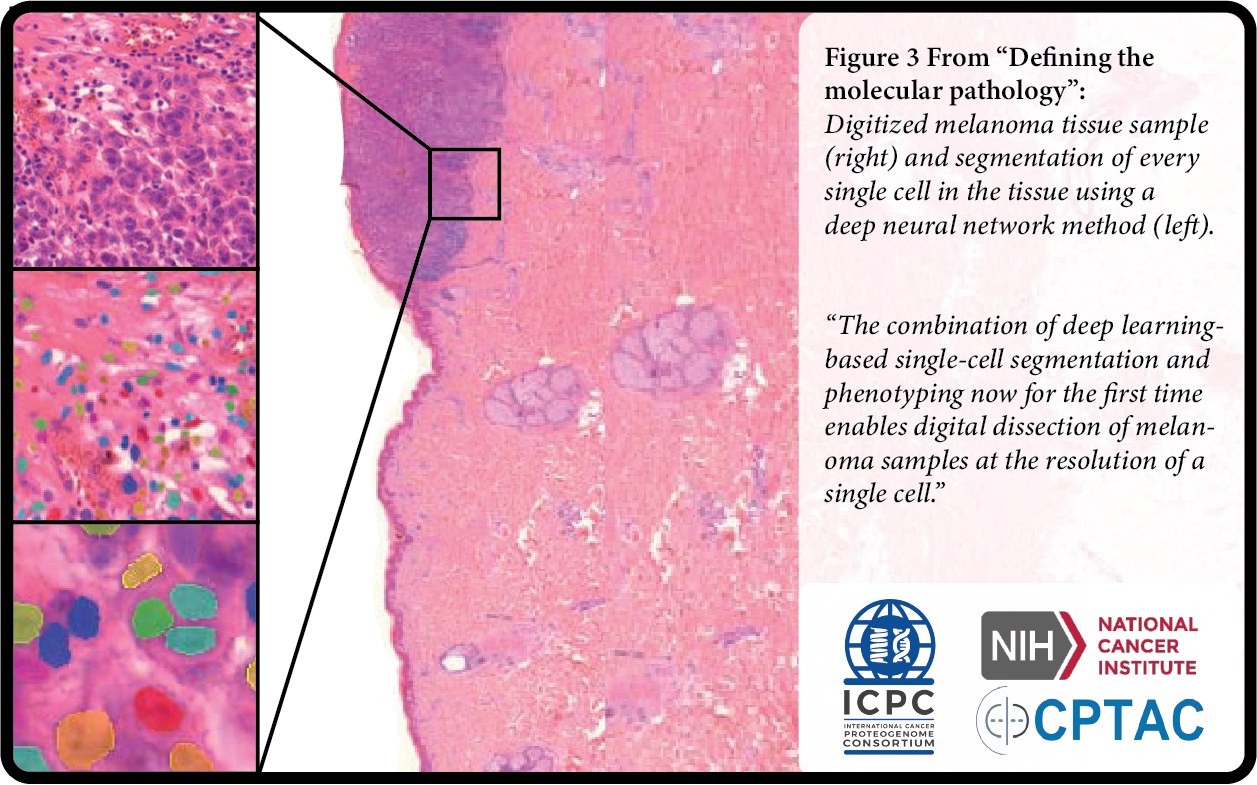
The two publications are integral parts of a series on the Human Melanoma Proteome published by Clinical and Translational Medicine. The first manuscript, “The Human Melanoma Proteome Atlas—Defining the Molecular Pathology,” describes the anatomical sites from which tumors were resected, the clinicohistopathological features of the MM500 cohort, a detailed histological characterization of the samples, and introduces the protein profiles of analyzed melanoma tumors. It also includes details of chromosomal and cellular localization, as well as differential expression of proteins in melanoma cultured cell lines and in tissues with high levels of tumor cells or stroma. The second publication is entitled “The Human Melanoma Proteome Atlas—Complementing the Melanoma Transcriptome”. This proteogenomic study integrates comprehensive proteomic analysis with genomic data from the NCI The Cancer Genome Atlas (TCGA) and establishes a knowledgebase mapped to the transcriptome and comprising expressed proteins, quantitation, spatiotemporal localization, mutations, splice isoforms, and PTM variants. It also describes the detection of proteins whose transcripts are missing from the TCGA transcriptome, melanoma driver mutations expressed at the protein level, and analysis of protein expression in the blood plasma of melanoma patients. Results from the combined body of work add extensive detail to the melanoma genome and its modifications with a direct bearing on protein function.
The MM500 cohort consisted of 505 samples from 294 tumors obtained from 232 patients. Among the identified 15,500 proteoforms, a large number of cytoplasmic proteins and 2,000 membrane proteins, such as receptors and ligands were included. The lab found that a notably high fraction of proteins identified were dedicated to cell division, splicing mechanisms, and metabolism. Additionally, results indicate that mitochondrial function is of particular importance in melanoma. The outline of the melanoma proteome, its integration with transcriptomic and mutation data, and combination with in-depth histopathological characterization of the tumors presented in the studies serves as a solid foundation for further discoveries in MM diagnosis and treatment. Dr. György Marko-Varga, one of the study’s leads, indicated that with support from the Berta Kamprad Foundation (IKEA), he is hopeful for the future of MM research, saying; “This [Proteome Atlas] is an important molecular pathology asset for the Global healthcare sector to utilize ‘for the benefit of cancer patients.”
This team is a proud member of the International Cancer Proteogenome Consortium – a National Cancer Institute (NCI) Cancer Moonshot-inspired initiative that provides a forum for collaboration among some of the world’s leading cancer and proteogenomic research centers.

