
Mark Baker's laboratory
at Macquarie University.
Credit: Samridhi Sharma
The Office of Cancer Clinical Proteomics Research at the National Cancer Institute, part of the United States National Institutes of Health, is spearheading the preparation and training of the proteogenomic research workforce on an international scale.
The NCI and Macquarie University in Australia, among other research agencies and institutes, are members of the International Cancer Proteogenome Consortium. Launched in 2016 and aligning efforts with the U.S. Cancer Moonshot Initiative, NCI and Macquarie University recently piloted an international student exchange in the field of proteogenomics—the comprehensive study of cancer patient tissue, proteins and genes to better understand cancer development, progression, diagnosis, therapeutic response, and potential novel therapeutic targets.
Memoranda of Understanding are foundational to ICPC. The MOU between NCI and Macquarie University, as well as other participating institutions, support training and the exchange of scientists with expertise in proteogenomic data. ICPC aims to improve patient outcomes by disseminating and sharing proteomics, genomics, and imaging data publicly with the international cancer research community.
Samridhi Sharma, a second-year doctoral student with Mark Baker, Ph.D., at Macquarie University, leveraged her postgraduate research fund to attend the 2017 Human Proteome Organization World Congress in Dublin, Ireland. She then traveled to the U.S. to complete four-weeks of proteogenomic training under Amanda Paulovich, M.D., Ph.D., a Clinical Proteomic Tumor Analysis Consortium Investigator and Member at the Fred Hutchinson Cancer Research Center in Seattle, Washington.
“I wanted to work in proteomics because it is a very powerful tool for potentially finding diagnostic plasma markers of early stage cancer,” Sharma said. “Proteomics is a strong technique that adds a high level of value to genomics, cell-based and animal model studies about particular biology or diseases, helping us filter out important protein candidates and moving forward with validation studies.”
Sharma is working to identify proteins in the blood that can act as diagnostic or prognostic markers for Stage I/II colorectal cancer. But before she can take the potential markers she has found to the clinic, Sharma had to learn how to develop laboratory methods to quantify the identified plasma proteins.
“In Dr. Paulovich’s lab, I learned how to do a technique called immuno-multiple reaction monitoring, or immuno-MRM, to pull down protein and peptides from plasma samples and then run [the samples] on a mass spectrometer,” Sharma said. “The objective of this is to validate our current identifiers from a prior SWATH study—the newest type of mass spectrometry—so that we can measure [the proteins] in multiple clinical samples.”
While studying the genome has led to new therapies, most of the current targeted cancer therapies pinpoint proteins. Dr. Paulovich and her research team have lead the development of the immuno-MRM technology platform to address the lack of reliable assays needed to measure human proteins, filling in some of the missing biology that has not been identified by genomically sequencing tumors alone.
“It made sense initially to focus on the genome. Then it became clear that a lot of patients seemed to not respond to drugs that they should respond to or they would respond initially and then develop resistance,” Dr. Paulovich said. “Our thought was if we looked at proteins, which are the targets of many new therapies, perhaps we could do a better job of predicting patient responses and understand the mechanisms through which the drugs work.”
According to Dr. Paulovich, the immuno-MRM technology platform is robust, precise, and can simultaneously measure multiple proteins in samples across a single run, a drastic improvement over the traditional platform. Immuno-MRM also uses an internal standard that can be implemented in the assay and used across laboratories, providing a standardized tool for quantifying human proteins with the end goal of improving patient diagnosis and treatment.
“Immuno-MRM is a technological capability that will help the translation of protein biomarkers into the clinical realm,” Dr. Paulovich said.
Dr. Baker, Professor of Proteomics and Biochemistry at Macquarie’s Faculty of Medicine and Health Sciences, says that the most important contribution to ICPC from Sharma’s work will be the knowledge to help us better understand the enemy that we know cancer to be.
“I hope that Samridhi’s work will show that we can see changes in the blood of patients affected with colorectal cancer at the very earliest stages when the cancer is no bigger than a pea,” Dr. Baker said. “These protein changes in the blood may reflect that the immune system recognizes the cancer as foreign in some way.”
As part of the international collaboration, the proteomics data that Sharma produces through her training will be contributed to the CPTAC Data Portal, which makes proteomics data publicly available for use by cancer researchers and physicians worldwide.
“We have agreed to participate in a data exchange common that aligns with the U.S. Cancer Moonshot Initiative, the NCI and CPTAC,” Dr. Baker said.
The data sets contributed by Dr. Baker and his research team fulfill a call-to-action in cancer research that represents the global diversity of people and commonly diagnosed cancers in their unique populations.
Dr. Baker’s expertise and contribution since working in the U.S. has made Macquarie University a center of proteomics and technology excellence, a hub for first-class proteomics research into colorectal cancer, and an obvious choice for collaboration.
Dr. Baker’s research group focuses on two simple aims that target colorectal cancer. The first aim is to find blood-based, early stage biomarkers that when combined with a conventional treatment like surgery can be used to cure cancer. The second aim focuses on developing drugs that reverse the biology of metastasis, a component of late stage cancer.
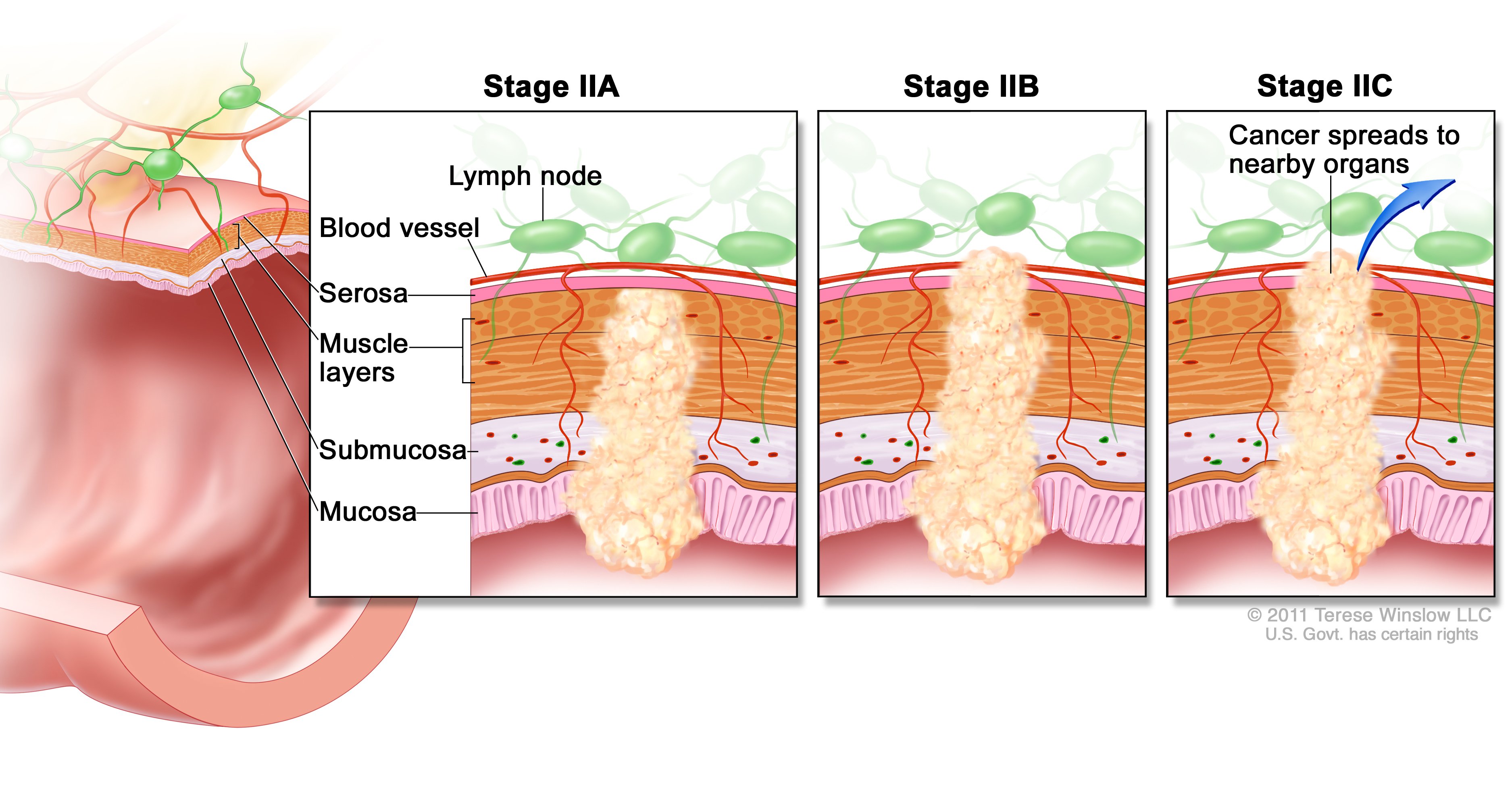
(left), cancer spreads through the
muscle layer of the colon/rectum wall to
the serosa. In Stage IIB (middle), cancer
spreads through the serosa but not to
nearby organs. Stage IIC (right), cancer
spread through serosa to nearby organs.
Credit: NCI Visuals
“We will detect a lot of early cancers that can be excised and save people’s lives. We want to get it before it recurs,” Dr. Baker said. “If [colorectal cancer] does recur, is it possible to slow it down and make it dormant for a little bit longer so we can prolong lives? To do that, we need to know the genes and proteins involved in cancer spread and how they work.”
According to the World Health Organization, colorectal cancer is the third most common cancer resulting in 1.36 million new cases and 694,000 deaths worldwide in 2012.
Australia has the highest incidence of colorectal cancer worldwide. While colon cancer can be a treatable and often curable disease, fewer than 40% of cancer cases are detected early enough according to Bowel Cancer Australia.
In 2015, Dr. Baker collaborated with Concord Repatriation General Hospital researchers to draw data from a 20-year colorectal cancer surveillance study. The teams discovered uPAR (urokinase plasminogen activator receptor), a potential biomarker that may allow clinicians to better predict whether a Stage II colorectal tumor is likely to recur or whether surgery has been successful.
According to Dr. Baker, Stage IIB patients have cancer that is thought to be confined within the bowel wall, not spreading to nearby organs or affecting the lymph nodes. Surgery is a treatment option for Stage II patients, however, 25% of patients on average still recur, leaving patients with a much lower survival rate. If uPAR is expressed on the cancer cells in the primary tumor, it may predict recurrence in Stage II patients.
“Clinicians in the past haven’t known how to judge if a Stage II patient is high risk, going to recur, or if surgery will be curative,” Dr. Baker said. “This could be a real personalized approach to this population of rectal cancer patients, affecting approximately 200,000 people on the planet every year.”
By using uPAR expression as a potential survival indicator in this subset of cancer patients, Dr. Baker hopes to arm clinicians with a simple test to further advise patients about the likelihood of cancer recurrence.
ICPC is fostering international collaborations like those between Drs. Baker and Paulovich laboratories, arming the international cancer research community with the knowledge necessary to better fight the terrible enemy that the world understands to be cancer.
“We’re starting to see new therapies, better detection of cancer earlier, and improvements to existing interventions based on the fact of knowledge,” Dr. Baker said. “The more knowledge you have about your enemy, the better off you are at fighting it.”

