Proteomic profiling often entails 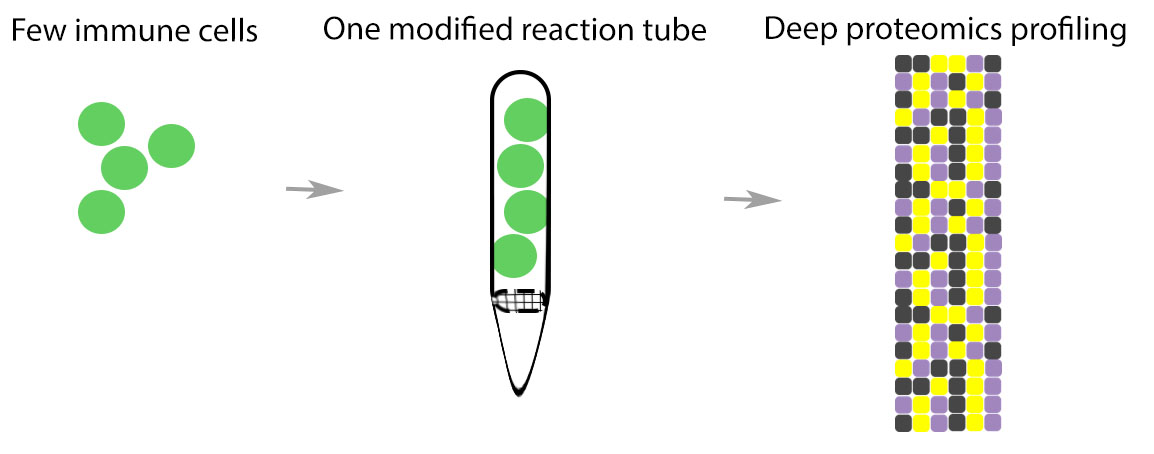 using a large sample input to explore the dynamic nature of protein expression and regulation. However, there are instances when researchers have the opposite due to prior methodology such as fluorescence activated cell sorting (FACS) or when dealing with patient samples. To overcome these technical challenges, CPTAC researchers at the Broad Institute have developed a technique (demonstrated on mouse immune cells) that uses such samples while achieving similar outputs of larger sample inputs.
using a large sample input to explore the dynamic nature of protein expression and regulation. However, there are instances when researchers have the opposite due to prior methodology such as fluorescence activated cell sorting (FACS) or when dealing with patient samples. To overcome these technical challenges, CPTAC researchers at the Broad Institute have developed a technique (demonstrated on mouse immune cells) that uses such samples while achieving similar outputs of larger sample inputs.
Immune cells are generally classified through transcriptional profiling which is looking at mRNA expression levels. Although the central dogma of biology describes the method by which information is transcribed from genes and translated into proteins, surprisingly, the proteome has yet to be thoroughly investigated. As cell loss is common in FACS, CPTAC researchers modified a pipette tip to allow FAC-sorted cells to be captured, lysed and digested in one reaction. This device, termed a micro-reactor, is then transferred to a second tube where the digested sample is then tagged for subsequent mass spectrometry analysis. Researchers used on-column tandem mass tag (TMT) labeling and multiplexing to bring multiple low input sample signals together to one stronger signal.
After this, researchers used statistical analysis to determine the reliability of their method, finding that hallmark signaling pathways were observed amongst the 12 immune cell types examined. Depth of coverage was still obtained; researchers identified over 7000 protein groups from their initial low input method. In fact, proteomic profiling classified additional markers that could be used to delineate cell types.
The true “biological insight” of this method, however, was when CPTAC researchers compared mRNA expression levels with protein expression levels. They found that co-expressed proteins do not always track with their respective co-expressed mRNA transcript levels, indicating that post-transcriptional regulation may play a role in innate immunity. Overall, this simple protocol uses a one-tube reaction to both allow low sample input and prevent cell loss while still maintaining robust signal and depth of coverage.
This article was published in Molecular & Cellular Proteomics.

