Protein ubiquitylation is best known for its role in degradation, leading molecules to the cell’s recycle bin, but it also has roles in cellular processing and protein interactions. Unfortunately, research using proteomic techniques have been limited due to experimental and technical complications.
Traditionally, SILAC (stable isotope labeling by amino acids in cell culture) 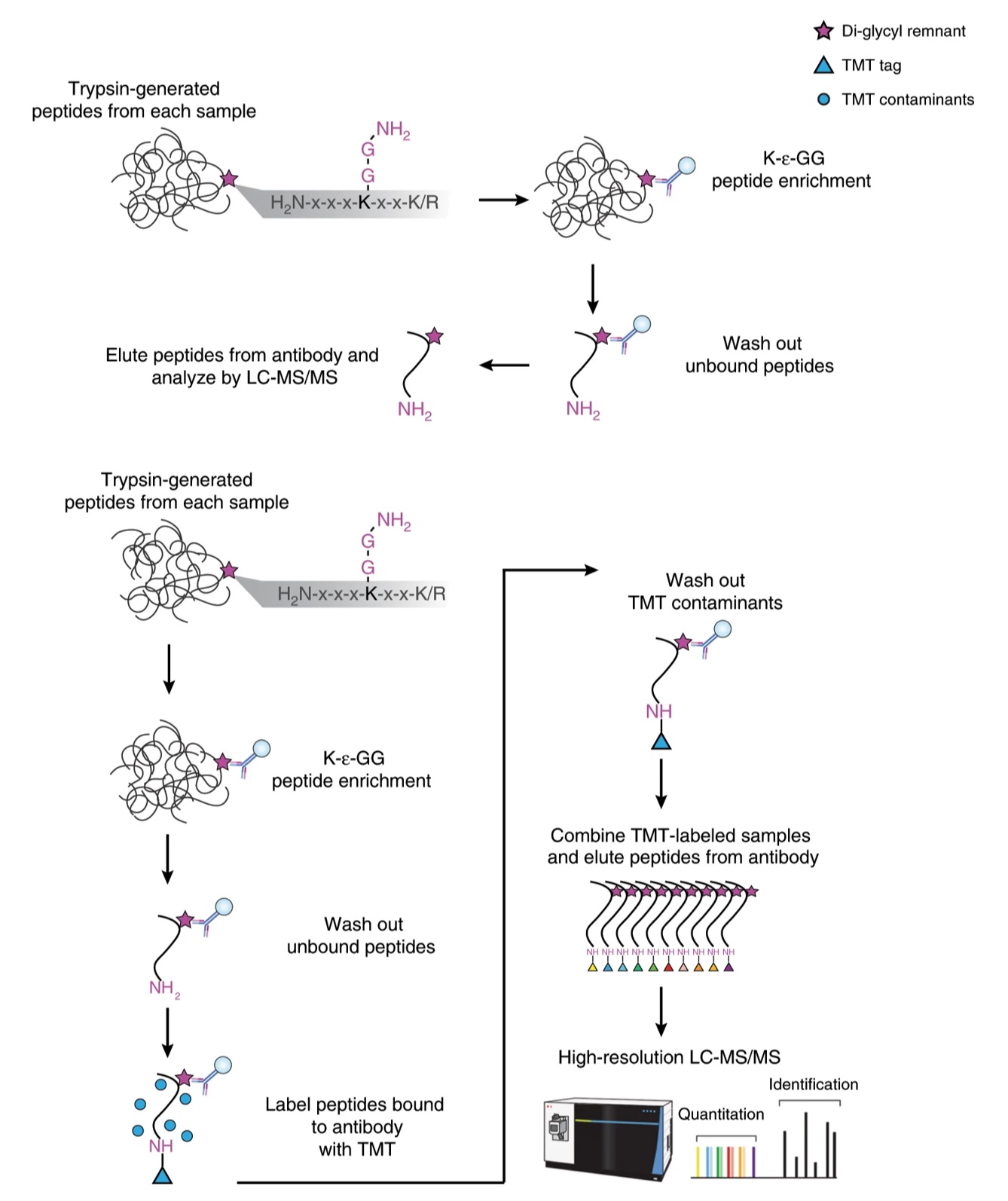 was the preferred method to proteomically profile ubiquitin. However, this method could not be applied to human or animal tissue and was limited to a 3-sample capacity per single experiment.
was the preferred method to proteomically profile ubiquitin. However, this method could not be applied to human or animal tissue and was limited to a 3-sample capacity per single experiment.
A recent article in Nature Communications describes how Clinical Proteomics Tumor Analysis Consortium (CPTAC) researchers developed a new way to look at ubiquitylation in tissue. This method, dubbed UbiFast, is designed to enable multiplexed, quantitative profiling of the ubiquitylome, using much smaller amounts of sample and substantially less instrumentation time than previous approaches. UbiFast takes a novel approach to isobaric chemical labeling of ubiquitin peptides with TMT reagents, solving several short comings of previous proteomic methods.
Isobaric chemical labeling, however, came with its own challenges. Commercially available antibodies do not specifically recognize or enrich ubiquitinylated peptides that have been labeled with TMT or iTRAQ. To circumvent this obstacle, CPTAC scientists from the Broad Institute of Harvard and MIT developed a method in which ubiquitin K-Ɛ-GG peptides were labeled with TMT reagents while still bound to anti-K-Ɛ-GG antibody beads prior to LC-MS/MS analysis. This allowed sample multiplexing, less TMT-based contamination (leading to greater assay sensitivity), and no cumbersome fractionation step – so a faster time to MS analysis. Scientists applied workflows from previous SILAC-based and TMT in-solution labeling proteomic studies to compare identification depth and mostly importantly quantitative accuracy. They were able to confirm protein targets of lenalidomide treatment, an anti-tumor drug for multiple myeloma, through changes in ubiquitylation of two transcription factors, Ikaros and Aiolos.
For proof of concept, Broad researchers applied this technique to basal and luminal breast cancer patient-derived xenograft (PDX) tissue samples. The experiment utilized 0.5mg of tissue (per sample) in a TMT 10-plex LC-MS/MS analysis that took a little over five hours and quantifying greater than 10,000 distinct ubiquitylation sites. Ubiquitylation plays an important role in immune signaling and, results described showed upregulated ubiquitylation of immune signaling proteins in the basal breast cancer model, consistent with the immune-hot nature of this cancer subtype relative to other breast cancer subtypes.
This study highlights the possibility of investigating the role of ubiquitylation in tumors and is currently being applied under the auspices of the CPTAC program. Furthermore, as exemplified in this study, this method highlights the possibility of investigating crosstalk between lysine acetylation and ubiquitylation.
In summary, the UbiFast method enables sensitive and efficient proteomic analysis of ubiquitylation, that can be multiplexed and potentially used for high-throughput analysis of tumor tissue as well as cells.

