A study published Feb 13th in Cell provides an unprecedented look at the dozens of molecular steps that occur to bring about endometrial cancer, commonly known as uterine cancer. The study offers insights about how physicians might be able to better identify which patients will need aggressive treatment and which won’t, and offers clues about why a common treatment is not effective with some patients.
The study, funded 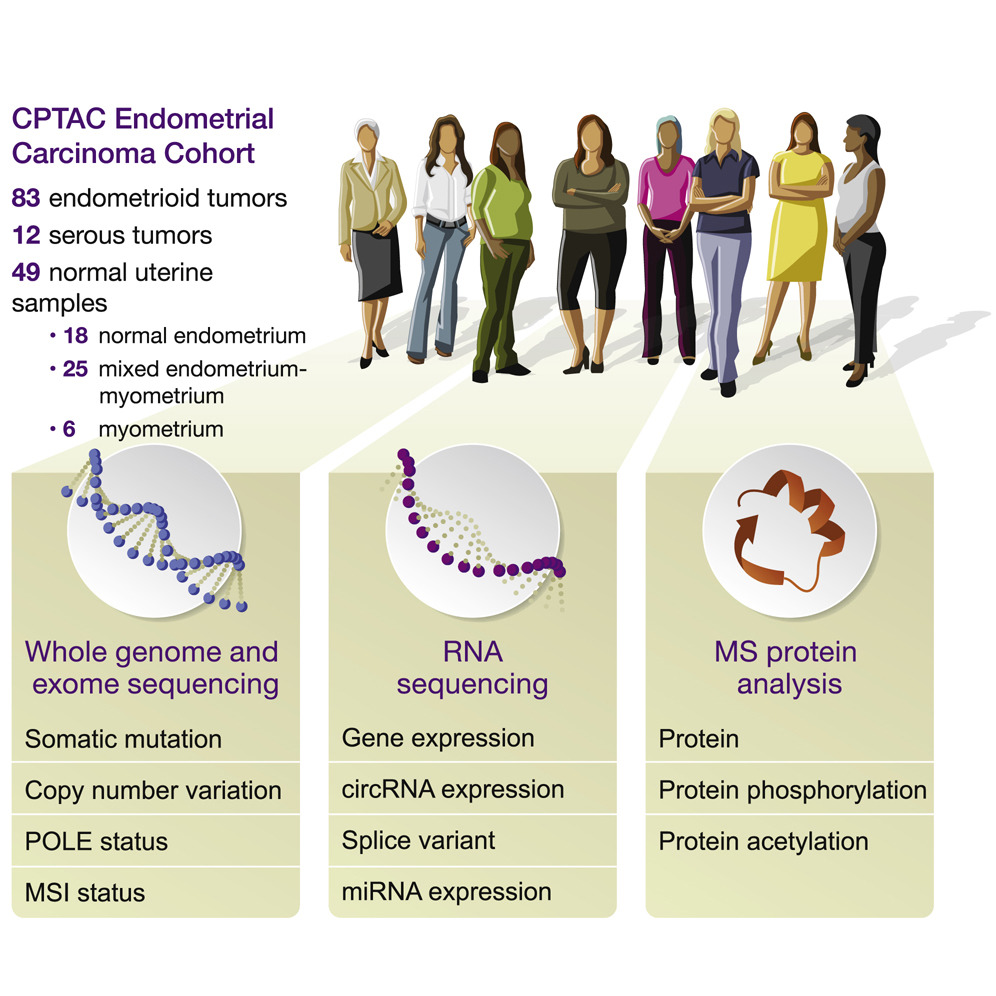 by the National Cancer Institute, also suggests a potential role for drugs that target proteins known as CDK12, SMARCA4 and PML in other types of cancer.
by the National Cancer Institute, also suggests a potential role for drugs that target proteins known as CDK12, SMARCA4 and PML in other types of cancer.
One could say that the work is a detailed molecular snapshot of endometrial cancer – except that the information is so vast, touching upon tens of thousands of molecular actors taking part in thousands of interactions at different times, that the work is more like a frame-by-frame video, documenting steps that play out over years in patients’ bodies.
“This is like the Google Earth of endometrial cancer,” said Karin Rodland, one of five corresponding authors of the paper and a cancer biologist at the Department of Energy’s Pacific Northwest National Laboratory. “It’s a very comprehensive portrait of this particular cancer type. We tried to measure everything we possibly could. Then we searched for patterns.”
Rodland and PNNL’s Tao Liu are two of five corresponding authors of the paper; the others come from New York University School of Medicine, Washington University in St. Louis, Baylor College of Medicine, and PNNL. Overall, scientists from more than a dozen institutions contributed. Senior author is David Fenyo of NYU.
The paper builds on work by The Cancer Genome Atlas, or TCGA, which identified the genetic underpinnings of the disease in 2013.
But genes are only the start of the story when it comes to cancer. When and where are those genes turned on, turned off, or mutated? What do they produce and how do those products interact?
The team studied 95 uterine tumors as well as 49 normal uterine tissue samples. Scientists measured the abundance, modifications and locations of a dizzying array of molecular players, including genes, messenger RNAs, circular RNAs, micro RNAs, and evidence of what scientists call “post-translational modifications.” These actions, including phosphorylation and acetylation, are key to determining when and where proteins, the molecular workhorses of every cell, are on or off.
Altogether the team took more than 12 million measurements.
What did they learn from the painstaking process? Here are the top takeaways.
Identifying the most aggressive tumors
Scientists developed a promising new way to identify tumors that currently are classified as not aggressive but which turn out to be just as invasive as “serous” tumors which grow quickly and are more likely than other tumors to kill patients.
Right now the aggressiveness of an endometrial tumor is determined largely by viewing the cells under a microscope. The team explored an additional method, showing how the activity levels of certain proteins clearly differentiate aggressive from less-aggressive tumors. As one example, the team showed how the protein beta-catenin, a well-known actor in many types of cancer, interacts with a signaling pathway known as Wnt to evade detection, accumulate and spur cells to grow out of control.
Unexpectedly, the team also found that a process that involves packing and unpacking genes happens more often than expected in tumor cells. With more than seven feet of DNA squeezed into nearly every one of our cells, the body packs it super efficiently, but it needs to unspool the DNA so that other molecular machinery can access it. The team’s measures indicate that a key part of this process, known as histone acetylation, is very active in endometrial cancer.
How cells become invasive
The team discovered that small, often-overlooked molecules known as circular RNAs seem to be involved in the transformation that cells undergo when they gain the ability to spread. Under normal circumstances, these circular RNAs will start a process to allow the cells to become mobile - the transition is called the “endothelial-mesenchymal transition” or EMT. In endometrial cancer, this regulatory checkpoint is turned ‘on’ and turned ‘up’, allowing the tumor cells to move around - it’s what makes endometrial cancer deadly.
Identifying patients who will, won’t benefit from ‘checkpoint therapy’
The team proposed a new way to determine which patients are most likely to benefit from a treatment known as checkpoint therapy, where drugs like pembrolizumab and nivolumab are used to dodge the barriers that some cancer cells use to evade the immune system. It’s one of many ways that physicians are exploring the use of “immunotherapy” to spur the body’s natural defenses to fight cancer. Better insight into exactly who would benefit from the drugs would allow physicians to avoid their use in patients who are unlikely to benefit, sparing those patients severe and unnecessary side effects.
Currently in endometrial cancer, physicians use a measurement known as “tumor mutation burden” to determine which patients are most likely to benefit. Based on the measurements of immune activity in this study, the scientists have proposed a new measure focused on a patient’s “antigen presentation machinery” or APM. The APM describes how well the body flags cancer cells and presents them to the body’s immune system for destruction – a key function for checkpoint inhibitors to be effective.
“This work contributes to the personalized medicine we need to deliver for patients who have endometrial cancer,” said Bing Zhang of Baylor College of Medicine, a corresponding author. “Such work will help us to know which patients will benefit most from which therapies.”
The study is the fifth publication in a series of studies funded by the National Cancer Institute’s Clinical Proteomic Tumor Analysis Consortium (CPTAC) to tackle the biological pathways involved in cancer. Previously published papers have focused on kidney, breast, ovarian and colon cancers.
This work was supported by grants U24CA210955, U24CA210954, U24CA210972, U24CA210979, U24CA210986 and U01CA214125 from the National Cancer Institute’s Clinical Proteomic Tumor Analysis Consortium (CPTAC), by grant RR160027 from the Cancer Prevention & Research Institutes of Texas (CPRIT), and by funding from the McNair Medical Institute at The Robert and Janice McNair Foundation. The measurements of proteins and protein activity were done at EMSL, the Environmental Molecular Sciences Laboratory, a user facility funded by the DOE’s Office of Science and located at PNNL.

