Protein complexes are a fundamental component of physiological and pathological processes. Despite organisms having a limited number of genes and therefore a finite number of proteins at their disposal, the number of possible combinations amongst these proteins is essentially inexhaustible; myriad protein complexes are constantly being assembled and disassembled at different physiology and pathological status in order to perform the complex and diverse biological functions. Characterization of these native protein complexes in vivo is critical to understanding their biological and clinical significance and is one of the major focuses of functional proteomics as a field. CPTAC researchers have developed an approach, recently published in Analytical Chemistry, to study protein complexes using in vivo cross-linking, followed by size exclusion chromatography (SEC) and data-independent acquisition mass spectrometry (DIA-MS).
At present, affinity purification mass spectrometry (AP-MS), protein co-fractionation using SEC coupled with mass spectrometry (Co-Frac-MS), and cross-linking mass spectrometry (XL-MS) are recognized as powerful approaches for the largescale analysis of protein complexes. However, challenges associated with determining which complexes formed in vivo prior to lysing the cells/tissues represent a bottleneck for these techniques. in vivo chemical cross-linking mass spectrometry circumvents this problem due to its ability to stabilize protein interactions in their native environment prior to cell lysis. In their publication, CPTAC researchers outline an integrated approach called X-Co-Frac-MS which combines the power of Co-Frac-MS methods with the desirable features of XL-MS before cells or tissues were lysed in order to characterize in vivo protein complexes accurately
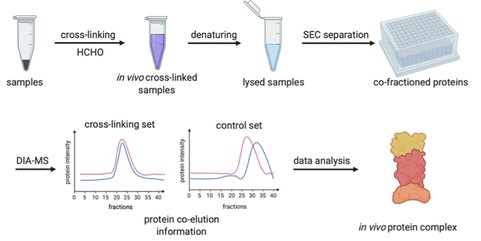 The X-Co-Frac-MS method consists of in vivo cross-linking, denaturing, SEC separation, DIA-MS, and data analysis steps. First, formaldehyde is used to cross-link and preserve in vivo protein complexes from intact cells or tissues. The team chose formaldehyde for in vivo cross-linking over other cross-linking reagents due to its small size, fast reaction, excellent cell and tissue permeability, broad utility in preserving tissues, as well as applicability in studying the interaction networks of protein complexes. Next, proteins from cross-linked cells are denatured in 8 M urea buffer to prevent in vitro protein complex formation and the denatured proteins and cross-linked protein complexes are then separated by SEC. The proteins collected from each SEC fraction are digested to peptides and analyzed by DIA-MS for protein identification and quantitation. The retention time of each protein is compared between the control and cross-linked fractions to determine in vivo protein complexes based on the protein retention time shift. The researchers tested the workflow and successfully applied it to characterize protein complexes and used criteria to confirm unique complexes. Additionally, to determine if a protein complex was formed directly through protein−protein interaction, XL-MS can be employed to identify protein interconnectivity at the residue level to distinguish direct from indirect interactions of protein complexes.
The X-Co-Frac-MS method consists of in vivo cross-linking, denaturing, SEC separation, DIA-MS, and data analysis steps. First, formaldehyde is used to cross-link and preserve in vivo protein complexes from intact cells or tissues. The team chose formaldehyde for in vivo cross-linking over other cross-linking reagents due to its small size, fast reaction, excellent cell and tissue permeability, broad utility in preserving tissues, as well as applicability in studying the interaction networks of protein complexes. Next, proteins from cross-linked cells are denatured in 8 M urea buffer to prevent in vitro protein complex formation and the denatured proteins and cross-linked protein complexes are then separated by SEC. The proteins collected from each SEC fraction are digested to peptides and analyzed by DIA-MS for protein identification and quantitation. The retention time of each protein is compared between the control and cross-linked fractions to determine in vivo protein complexes based on the protein retention time shift. The researchers tested the workflow and successfully applied it to characterize protein complexes and used criteria to confirm unique complexes. Additionally, to determine if a protein complex was formed directly through protein−protein interaction, XL-MS can be employed to identify protein interconnectivity at the residue level to distinguish direct from indirect interactions of protein complexes.
Due to the bulk of existing data containing protein complexes being collected via methods that don’t preserve the in vivo state of the cells/tissues, this new method should provide a wealth of unique information on authentic protein complexes which occur in living systems. Importantly, this method can be generalized for characterizing in vivo protein complexes from various sample types, including tissues, for both research and clinical applications in the future.

