Despite advances in the efficacy and specificity of cancer treatments over the last decade, pancreatic cancer remains one of the leading causes of death worldwide. The most common type of pancreatic tumor, Pancreatic Ductal Adenocarcinoma (PDAC), has distinctly poor patient outcomes due to its aggressive progression and late symptom presentation.
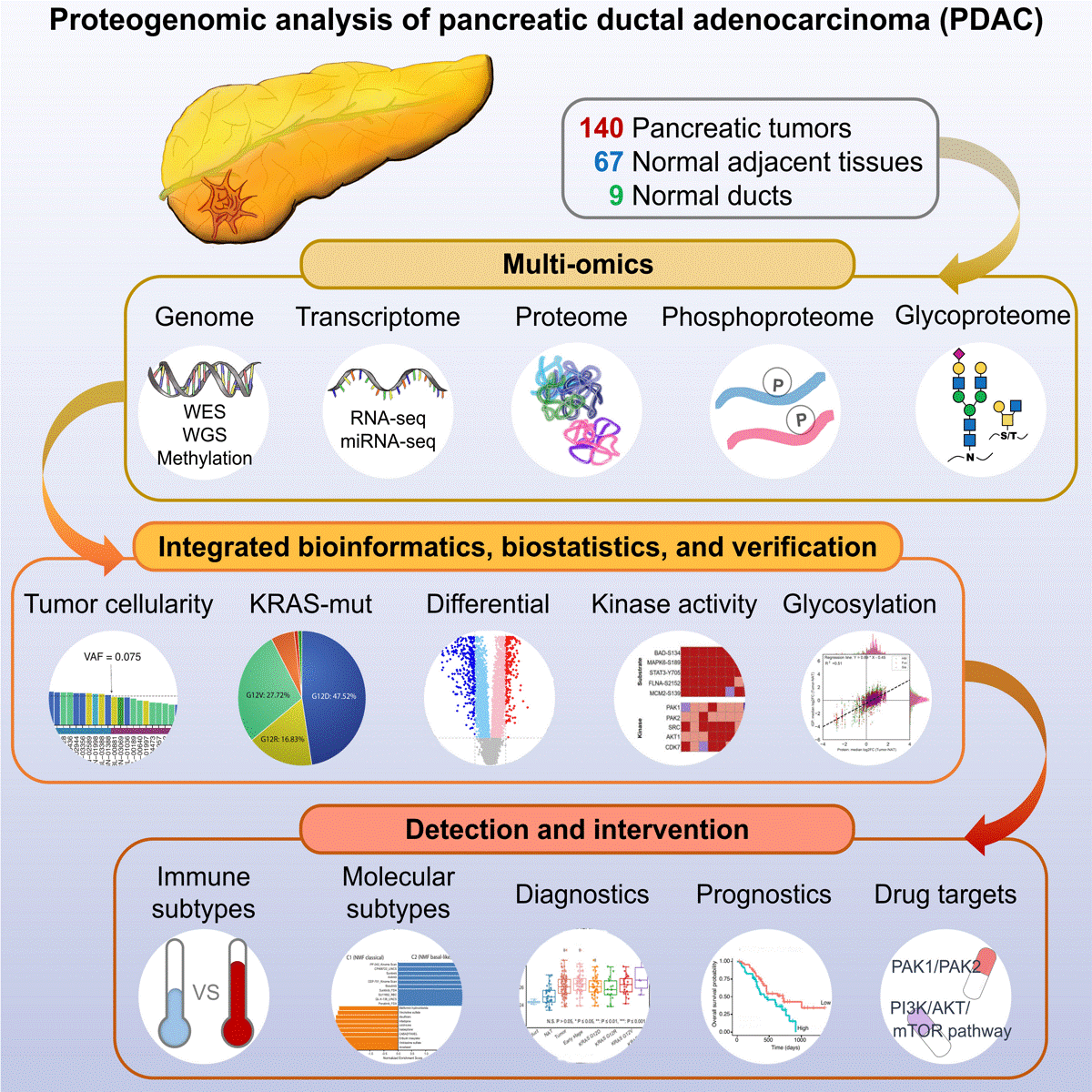 To learn more about biology and identify new molecular features of pancreatic cancer that can be harnessed for early detection and treatment, the Clinical Proteomic Tumor Analysis Consortium (CPTAC) undertook a large-scale, international study led by investigators from Johns Hopkins University, Baylor College of Medicine, Washington University School of Medicine in St. Louis, and University of Calgary. This collaborative effort, described in Cell, brings proteomic, transcriptomic, and genomic data together into a detailed "proteogenomic" view of PDAC. Study findings could offer hope to patients with this deadly disease. Data (genomics, proteomics, radiology, and pathology images) from the study can be accessed at the National Cancer Institute’s Proteomic Data Commons.
To learn more about biology and identify new molecular features of pancreatic cancer that can be harnessed for early detection and treatment, the Clinical Proteomic Tumor Analysis Consortium (CPTAC) undertook a large-scale, international study led by investigators from Johns Hopkins University, Baylor College of Medicine, Washington University School of Medicine in St. Louis, and University of Calgary. This collaborative effort, described in Cell, brings proteomic, transcriptomic, and genomic data together into a detailed "proteogenomic" view of PDAC. Study findings could offer hope to patients with this deadly disease. Data (genomics, proteomics, radiology, and pathology images) from the study can be accessed at the National Cancer Institute’s Proteomic Data Commons.
Searching for new ways to fight pancreatic cancer, the team took a new look at pancreatic tumors from several different angles. They compared 140 tumor samples, 67 samples of normal adjacent pancreatic tissue, and 9 samples of pancreatic tissue from non-cancerous patients. Utilizing a thorough proteogenomic approach, the team looked at the entire genome and a plethora of relevant proteins.
“Although numerous studies have examined the genomics of pancreatic tumors and identified several mutations linked to this disease,” noted study leader Hui Zhang, Ph.D., M.S., from Johns Hopkins University, “drug therapies to target these mutations have been unsuccessful.” In addition, these tumors also don’t attract a significant response from the immune system, so immunotherapies have not been broadly effective. Also, Oliver Bathe, M.D., M.Sc., FRCSC, FACS, from the Arnie Charbonneau Cancer Institute at the University of Calgary, noted that "this very significant advance would only have been possible with the cooperation of a highly dedicated and learned group of investigators from a wide variety of disciplines from many institutions."
This research confirmed that pancreatic tumors are more likely to have mutations in several genes identified in previous studies, including KRAS, TP53, CDKN2A, and SMAD4. Unique to this study, comprehensive proteomic analysis identified 222 proteins with at least a two-fold increase in abundance between pancreatic cancerous cells and normal cells; nearly 5,000 sites in these proteins with increased phosphorylation abundance; and more than 1,700 sites with increased glycosylation abundance. “Several of these glycosylated proteins are secreted from pancreatic cancer cells,” says Zhang, “suggesting that they could potentially be captured in the blood for early diagnosis."
Other protein differences between the cancer cells and normal tissue - including multiple kinases such as PAK1 and PAK2-- suggest new opportunities to target pancreatic cancer. “Positioned downstream of KRAS, which is mutated in more than 95% of pancreatic tumors but remains largely untargetable, PAKs are attractive targets for further investigation,” noted Bing Zhang, Ph.D., from Baylor College of Medicine. Furthermore, Li Ding, Ph.D., from Washington University School of Medicine in St. Louis, noted that “over two dozen of proteins have been found to be upregulated in pancreatic tumors. Among them, increased LOXL2 protein level is tied to poor survival, potentially serving as a prognostic marker for this disease."
Overall, by studying these tissues using a proteogenomic approach, this study was able to confirm multiple clinically relevant biomarkers and further document the phenotypic effects of common genomic and epigenetic perturbations. Furthermore, and perhaps most importantly, the dataset from this study represents an invaluable resource for future studies focused on early detection and pancreatic tumor classification.
Source
Liwei Cao, Chen Huang, Daniel Cui Zhou, Yingwei Hu, et al. Proteogenomic characterization of pancreatic ductal adenocarcinoma. Published online September 17, 2021. DOI: 10.1016/j.cell.2021.08.023

