The following is the transcript of an interview conducted to highlight promising undergraduate student researcher Aniket Dagar from University of Michigan. Aniket was recently awarded a meritorious poster award for the poster he presented at the 2022 American Association for Cancer Research (AACR) annual meeting titled “Discovery of a Candidate Prognostic Biomarker for Clear Cell Renal Cell Carcinoma based on Transcriptomic and Proteomic Analyses.”
How would you describe the work you do and what your AACR poster covered to someone who is not familiar with it or does not have a background in biology?
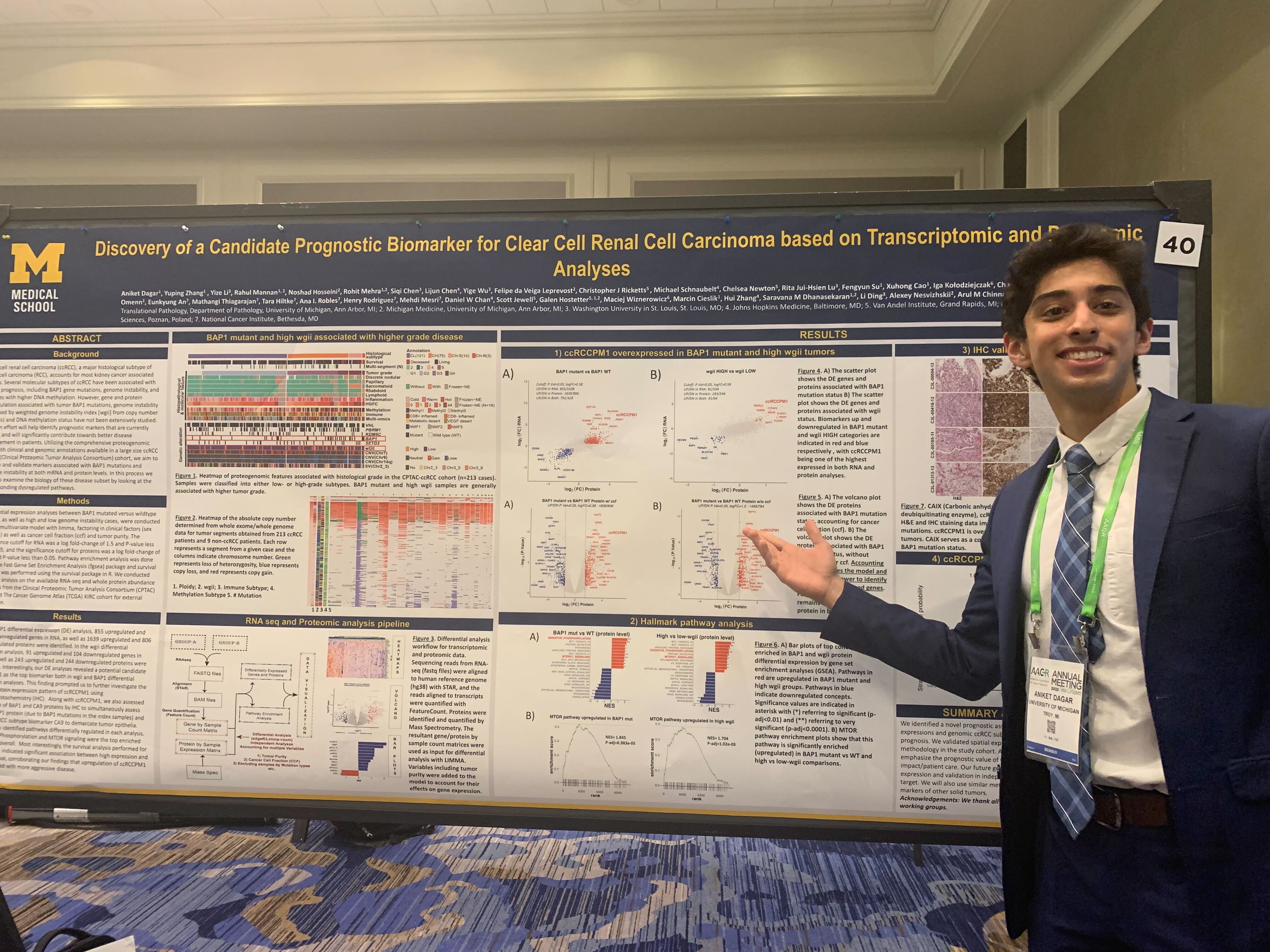
In cancer, multiple genes drive tumor growth. If a certain gene is upregulated, which means certain gene is expressed more, in cancer this might lead to a more aggressive cancer subtype. This is the focal point of my research. The genes involved in a specific subtype of kidney cancer called clear-cell renal cell carcinoma or CCRCC are responsible for a majority of kidney cancer deaths and is a very aggressive subtype of kidney cancer. So, this research investigates the biomarkers, or the genes, that are upregulated in CCRCC. For the research featured on the poster, I identified genes that were associated with more aggressive disease. I focused on two of the subtypes of CCRCC, BAP1 mutations and high weighted genome instability (High wGII). Both subtypes lead to more aggressive disease. BAP1 mutations are basically gene mutations in a chromatin remodeling gene which leads to cell instability and proliferation. High wGII occurs when the chromosome copy number variation is higher than the median ploidy. An analysis was performed comparing the BAP1 tumors versus the BAP1 wild-type and then the high wGII versus the low wGII and from this analysis, very interestingly, 1 biomarker, one gene was found to be upregulated in both the BAP1 mutant and the high wGII. So, in both very aggressive CCRCC diseases, the same gene was found to be upregulated. And this kind of brought about the question: does this gene contribute to the aggressiveness of the cancer somehow? From there I performed a survival analysis and, looking at patients with high expression of CCRCCPM1 versus patients with low expression of it, the high expression group had much worse survival. This was significant to a very low P value, so this did not happen by chance. The clinical significance of this is that, with further study, validations, and different data sets, we may be able to use this gene as a marker in a clinical setting. So, when patients come in and we find that this gene is more upregulated in their cancer, we can predict that the disease will be more aggressive and tailor treatments for disease accordingly.
Where does the project start and end? Or is it an ongoing project? What are the future plans for the lab?
So we are investigating the CCRCC disease in our lab as well as with the National Cancer Institute's Clinical Proteomic Tumor Analysis Consortium (CPTAC) as part of their Kidney Cancer Working Group. Although we have published our findings in Cell this is still an ongoing project. In our lab we are trying to further characterize CCRCC, be that through validations or functional tests that show the CCRCC subtype is related to worse survival in patients. The work featured on my poster was kind of the big finding of my four years of undergraduate research. I've been studying kidney and prostate cancer, focusing on very small [features] of the disease. Kidney cancer and then the CCRCC subtype, so I've been focused on CCRCC for the majority of my four years.
After working with the same type of cancer for so long, what keeps you coming back to the research?
The main thing that really keeps involved is being able to expand my knowledge and making these discoveries that are directly relevant to patient care. I've realized that every month I become even more interested--even more invested in what I'm doing and the type of research that I do. Coding, reading papers online, improving my knowledge base. This is what keeps me interested. The more research I do on one specific topic, the more interest I gain, and the more research I want to do, so I guess it's kind of a cycle for me.
That's cool! I have two follow-up questions to that point. Does coding encompass the majority of the work you do in the lab?
Most of my research right now is focused on coding and I love that. When I started doing research in my junior year of high school through my freshman year of college, I was doing wet lab research and I did like that, too. I like having the hands-on experience. What really changed my focus was that, during the COVID quarantine summer my PI, Dr. Udager encouraged me to try coding and analysis in R. I had no programming experience at that time, and I spent most of the summer just learning it and this led me to the lab where I work now. My mentors, Dr. Mohan Dhanasekaran and Dr. Yuping Zhang, have really helped me improve my skills in R. Yuping has been my mentor [for] the coding aspect and Mohan is a mentor for the cancer biology. Learning it and working at it, I have become much more proficient with code and I really, really love what I'm doing.
Going off of that, how important do you think it is for young professionals in biology or related fields to get involved with computer science? It sounds like it has essentially shaped your research experience.
I would say I strongly recommend every high school or college student to at least try the programming and the analysis aspect of research. The future of research is going to be heavily based on bioinformatics. With sequencing costs dropping and availability of next-generation sequencing, nanopore sequencing, and all these other high-throughput methods which can be used to evaluate and sequence patient’s tumors, we will need bioinformaticians in the future to help out with these deep analyses and make new discoveries. I truly think the direction of the future is in sequencing, proteomics, and bioinformatics analysis. We can already see a trend heading towards that, even in the last five to 10 years. I would strongly recommend young professionals and younger undergraduates in college to give it a shot and, although it may not be hands-on learning, you really get an understanding of depth in the research you're doing, and you're seeing what results [your] analyses produce and how they contribute to the discovery of biomarkers.
Thanks for sharing that. What role would you say CTPAC plays in terms of the research your lab does and the work specifically featured on your AACR poster?
Yes, so CPTAC has probably been the most essential aspect of our research, and for my poster. So CPTAC’s extensive proteogenomic data-- the cohort that I analyzed was a 200-plus patient cohort-- with a large amount of proteomic data compiled, along with clinical and genomic annotations, has allowed us to do a more robust analysis. This is usually not possible with smaller data sets, so we had many more samples and lots of annotations that could be used for our analysis. I also just want to thank the CPTAC leadership for all their direction and guidance throughout the whole project and the entire kidney cancer working group, along with of course Drs. Alexey Nesvizhskii, Li Ding, Li Zhang, and my PI Dr. Chinnaiyan.
What is the trajectory you see your career going? Has the work you’ve done during your undergraduate research influenced this path?
That’s a great question. So when I first started research in high school, I wasn't 100% sure if I wanted to continue with it. What really changed my trajectory was my research at MACP (Michigan Center of Translational Pathology). My mentors Mohan and Yuping have been a crucial part of my learning there. Their diligence. Their hard work. The late nights at 2:00 AM on weekends. All of that has really rubbed off on me and helped me become a better researcher. For example, Mohan, he would schedule calls with me to discuss the biology of kidney cancer, my analysis, and my research. We would spend two to three hours discussing and from those calls I feel like I've learned so much more about my research and related research in the field. [My mentor] Yuping is one of the most hardworking people I know and one of my biggest role models in the lab. Her attitude in terms of getting the job done and always responding to me within minutes or teaching me the statistical analysis that has been really important to my learning. She is always there for me. For me to have [mentors] to look up to like that has played a crucial role in putting in me a fire to continue researching, continue learning, and continue making such discoveries. I definitely want to, in the future, want to attend medical school, and continue to combine my clinical skills with my informatics research skills for better personalized medicine.
Is there anything else you would like to highlight? Experiences that have shaped you as a researcher, scientist, and person?
My volunteer experience has also been of the highlights of my undergraduate experience. I’m the hospital tutoring director for OEC, which is Opportunities to Educate Children. I manage around 100 volunteers at Michigan Medicine to help provide educational support for kids in the hospital--so that means kids in a pediatric dialysis or pediatric oncology treatment. These kids miss weeks of school, sometimes months of school because they’re in the hospital and our job as tutors is to not only provide support as a tutor, but also as a friend. Someone who can be there for them during that difficult time. Another extracurricular that I'm very involved in is UMURJ, which is the undergraduate Michigan Research Journal, and I'm the editor in chief of that. This has been an important complement to my research because my research for the last 6-7 years has been exclusively about cancer. Through this activity I’ve been able to read papers in the humanities, in the arts, the social sciences, to broaden my understanding of the research world as a whole while developing my leadership skills as an editor in chief.
*Transcript Edited for Clarity*

