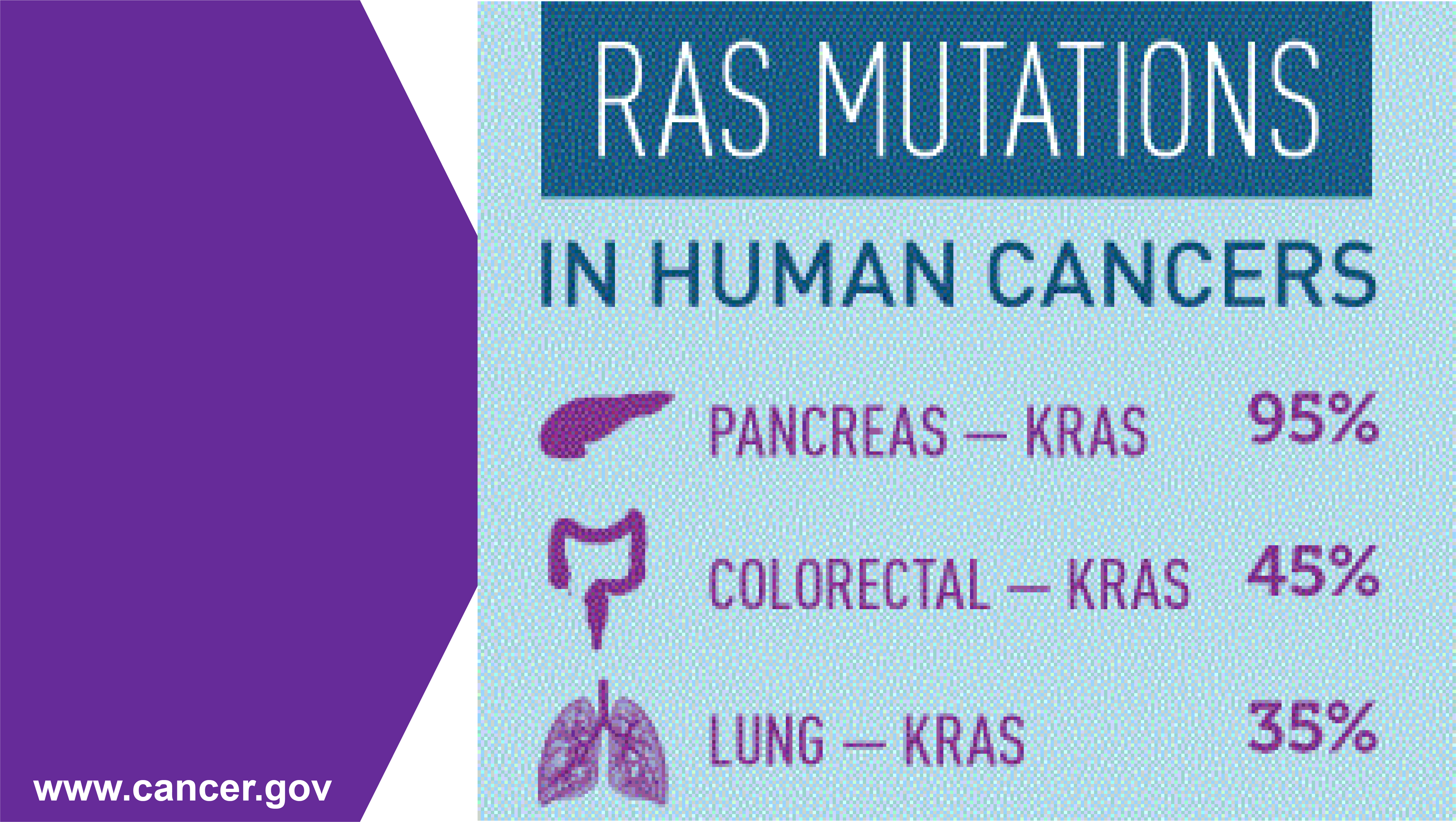
 Mutations in the RAS genes — KRAS, HRAS, and NRAS — have been identified in approximately 30% of all human cancers. While RAS gene family members encode proteins that are pivotal for cytoplasmic cell signaling, RAS oncogenes allow cells to grow uncontrollably and evade death signals. KRAS gene mutations are found at a high frequency in pancreatic (95%), colorectal (45%), and lung (35%) cancers, and chronic activation of its protein products can drive cancer progression and negatively impact treatment outcomes. Yet, key regulatory mechanisms involving KRAS isoforms 4A, 4B and their influence on pathological conditions such as cancer, particularly at the protein level, remain unclear. Dr. Neil Kelleher and his research team at Northwestern University used a targeted top-down (TD) proteomic workflow to detect distinct post-translational modification (PTM) profiles on intact KRAS4B isolated from both human colorectal cancer cell lines and tumor samples with KRAS gene mutations.
Mutations in the RAS genes — KRAS, HRAS, and NRAS — have been identified in approximately 30% of all human cancers. While RAS gene family members encode proteins that are pivotal for cytoplasmic cell signaling, RAS oncogenes allow cells to grow uncontrollably and evade death signals. KRAS gene mutations are found at a high frequency in pancreatic (95%), colorectal (45%), and lung (35%) cancers, and chronic activation of its protein products can drive cancer progression and negatively impact treatment outcomes. Yet, key regulatory mechanisms involving KRAS isoforms 4A, 4B and their influence on pathological conditions such as cancer, particularly at the protein level, remain unclear. Dr. Neil Kelleher and his research team at Northwestern University used a targeted top-down (TD) proteomic workflow to detect distinct post-translational modification (PTM) profiles on intact KRAS4B isolated from both human colorectal cancer cell lines and tumor samples with KRAS gene mutations.
As reported in the journal PNAS, the investigators combined pan-RAS immunoprecipitation (IP) with the TD approach to enrich and directly measure modified KRAS4B proteoforms with high molecular specificity, followed by a proof-of-principle experiment to optimize and validate the IP-TD assays in colorectal cancer cell lines harboring the KRAS G13D mutation. The proof-of-principle experiment detected a novel KRAS4B nitrosylation that could potentially support proliferation and survival in colorectal cancer cells bearing a single wild-type KRAS allele. When applying the IP-TD approach to deidentified colorectal tumors from the National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC) Biospecimen Core Resource, the investigators identified high variability in the degree of KRAS4B C-terminal carboxymethyation, a PTM critical for membrane association and productive RAS-mediated signaling, which may influence disease state or progression. These results have the potential to translate to clinical application and influence mutation-specific anti-RAS therapeutic strategies.
About the National Cancer Institute CPTAC: CPTAC is a national effort to integrate proteomic findings with genomics to accelerate the understanding of the molecular basis of cancer. CPTAC transforms tissue samples into publicly available proteomic datasets, assays, and antibodies for the cancer research community in efforts to expedite the development of individualized patient care.
About Frederick National Laboratory for Cancer Research: The Frederick National Laboratory for Cancer Research (FNLCR) Ras Initiative works to increase the sharing of knowledge and resources that are essential to defeat cancers caused by mutant RAS genes. RAS Initiative researchers develop unique reagents like protein standards and cell lines at FNLCR and widely distributes resources to the larger RAS community.

