The Clinical Proteomic Tumor Analysis Consortium (CPTAC) is pleased to release its newest comprehensive dataset - deep proteomic/phosphoproteomic data and imaging data of Lung Adenocarcinoma (LUAD) patient tumors. The CPTAC Tumor Characterization Program uses proteogenomic analysis to systematically identify proteins that derive from alterations in cancer genomes and related biological processes and provide this data with accompanying assays, reagents and protocols to the public that allows a wider group of scientists to extend and accelerate knowledge in unanticipated directions.
Lung adenocarcinomas make up about 40% of all lung cancers and primarily occurs in current or former smokers, but is the most common lung cancer among non-smokers. It is a leading cause of cancer-related mortality with more than a million deaths each year. The LUAD unique cohort boasts over 100 cases with Chinese and Vietnamese subpopulations comprising of about 50%, and Eastern European and American subpopulations making up the remaining half. Additionally, the study includes both males and females, and almost equal amounts of smokers and non-smokers. The prospectively collected, treatment naïve LUAD samples paired with adjacent histopathologically normal tissues, includes analysis for proteomics, phosphoproteomics, whole genome and exome sequencing, RNA-seq, DNA methylation and images. This data represents one of the most comprehensive multi-omics (DNA, RNA, protein, and imaging) datasets of LUAD patient samples in the world.
This completed dataset joins the deep comprehensive proteogenomic characterization of both Uterine Corpus Endometrial Carcinoma (UCEC) and Clear Cell Renal Cell Carcinoma (ccRCC) discovery datasets released in October 2018, also through the CPTAC Tumor Characterization Program. The rigorous attention given by CPTAC investigators from sample collection to proteogenomics profiling have produced a plethora of information for each of these studies. We are excited to have scientists use this publicly available information to target cancers for translational medicine.
Visit the CPTAC Data Portal to accept the data user agreement for file access. The CPTAC publication embargo ends January 1, 2020 for LUAD discovery data.
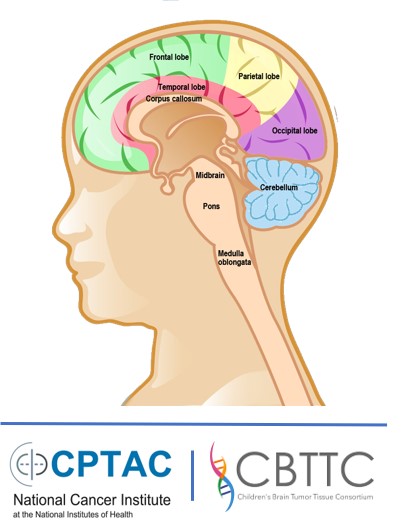 the same, there are more than 120 subtypes of brain cancers with very diverse features and diagnosis.
the same, there are more than 120 subtypes of brain cancers with very diverse features and diagnosis.
 Meeting of the Japanese Society of Nephrology
Meeting of the Japanese Society of Nephrology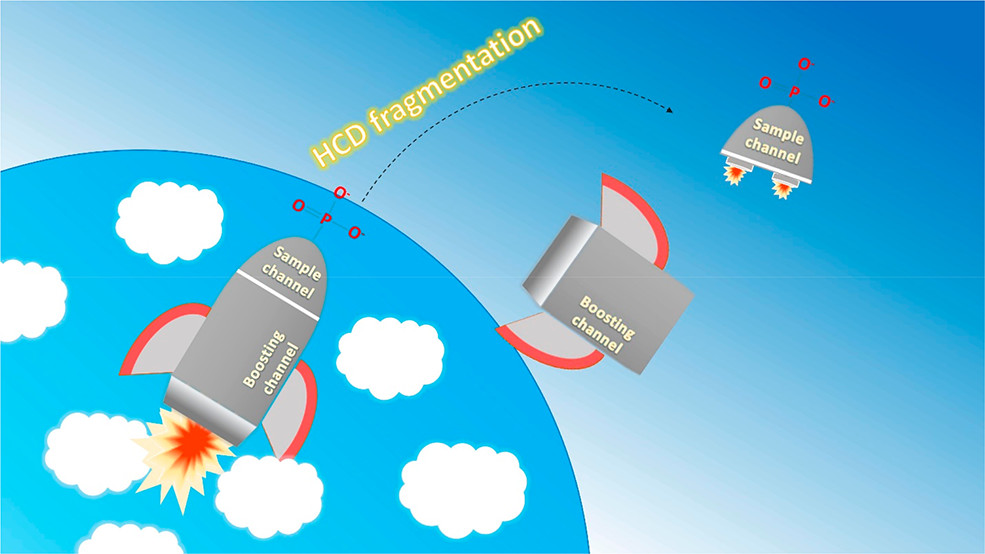 and has long been appreciated as an essential mechanism for the control of cellular function - tells a protein where to go, what to bind to and even when to die.
and has long been appreciated as an essential mechanism for the control of cellular function - tells a protein where to go, what to bind to and even when to die.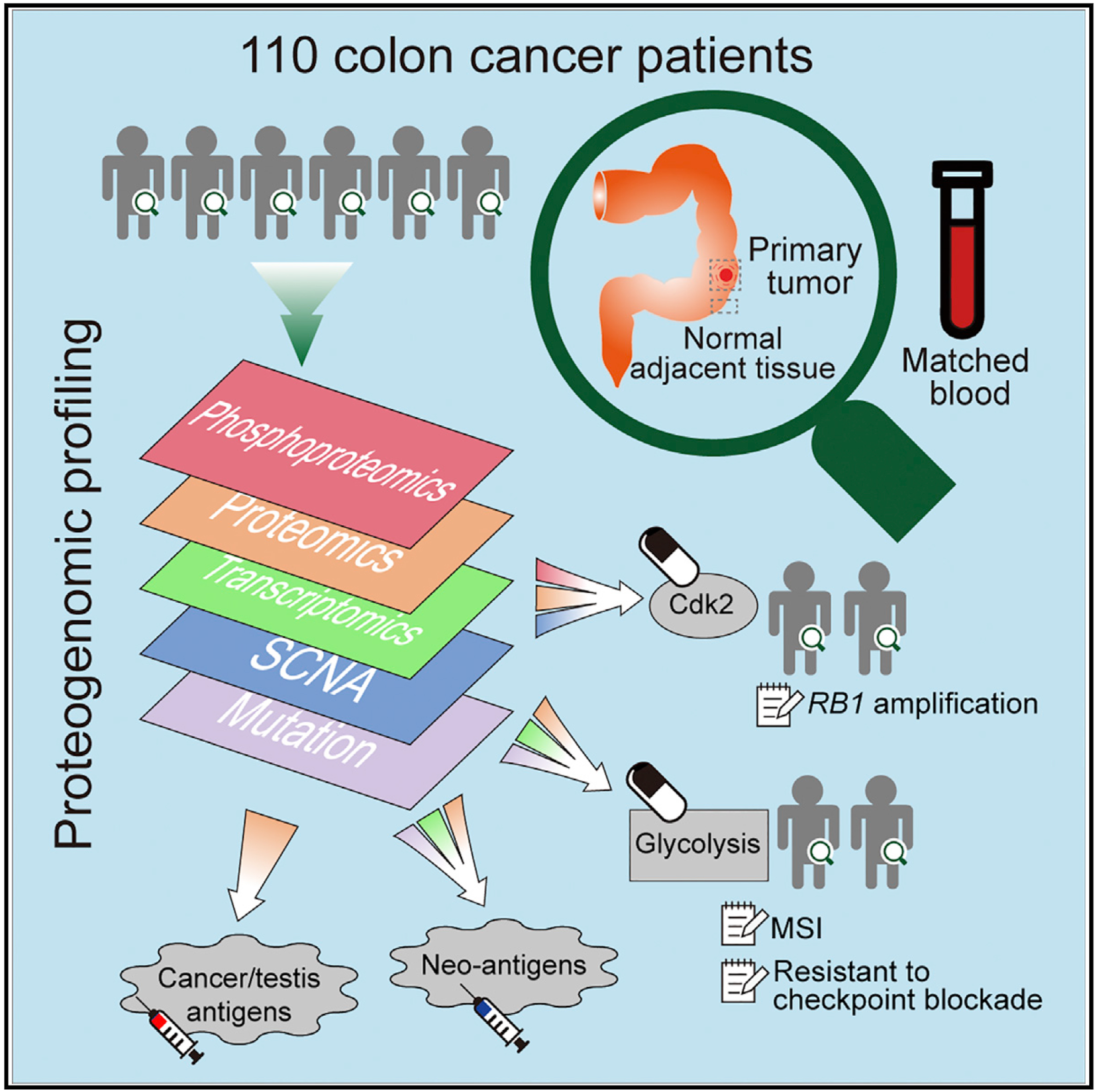 Analyzing both the entire set of genes and all the proteins produced by colon cancer tissues from patient samples has revealed a more comprehensive view of the tumor that points at novel cancer biological mechanisms and possible new therapeutic strategies.
Analyzing both the entire set of genes and all the proteins produced by colon cancer tissues from patient samples has revealed a more comprehensive view of the tumor that points at novel cancer biological mechanisms and possible new therapeutic strategies. Is DNA sequencing enough to recommend personalized treatments for cancer patients? In a article published in Nature Reviews Clinical Oncology, CPTAC investigators and colleagues from the Fred Hutch, Baylor College of Medicine, and University of Washington Medicine make the case for proteogenomics - analysis of the genomic and proteomic changes in cancer tumors.
Is DNA sequencing enough to recommend personalized treatments for cancer patients? In a article published in Nature Reviews Clinical Oncology, CPTAC investigators and colleagues from the Fred Hutch, Baylor College of Medicine, and University of Washington Medicine make the case for proteogenomics - analysis of the genomic and proteomic changes in cancer tumors. CPTAC is a comprehensive and coordinated effort to accelerate the understanding of the molecular basis of cancer through the application of robust, quantitative, proteomic technologies and workflows. The overarching goal of CPTAC is to improve our ability to diagnose, treat and prevent cancer.
CPTAC is a comprehensive and coordinated effort to accelerate the understanding of the molecular basis of cancer through the application of robust, quantitative, proteomic technologies and workflows. The overarching goal of CPTAC is to improve our ability to diagnose, treat and prevent cancer. "Big data" is a term to describe data sets so large or complex that traditional data processing strategies are inadequate. As continued advancements in biomedical technologies generate an increasing amount of patient data, administration of patient-centered care will depend, in part, on the ability to harness relevant insights from this data.
"Big data" is a term to describe data sets so large or complex that traditional data processing strategies are inadequate. As continued advancements in biomedical technologies generate an increasing amount of patient data, administration of patient-centered care will depend, in part, on the ability to harness relevant insights from this data.