ICPC China Team Comprehensively Characterizes HBV Related Liver Cancer
Hepatocellular carcinoma (HCC), the most common 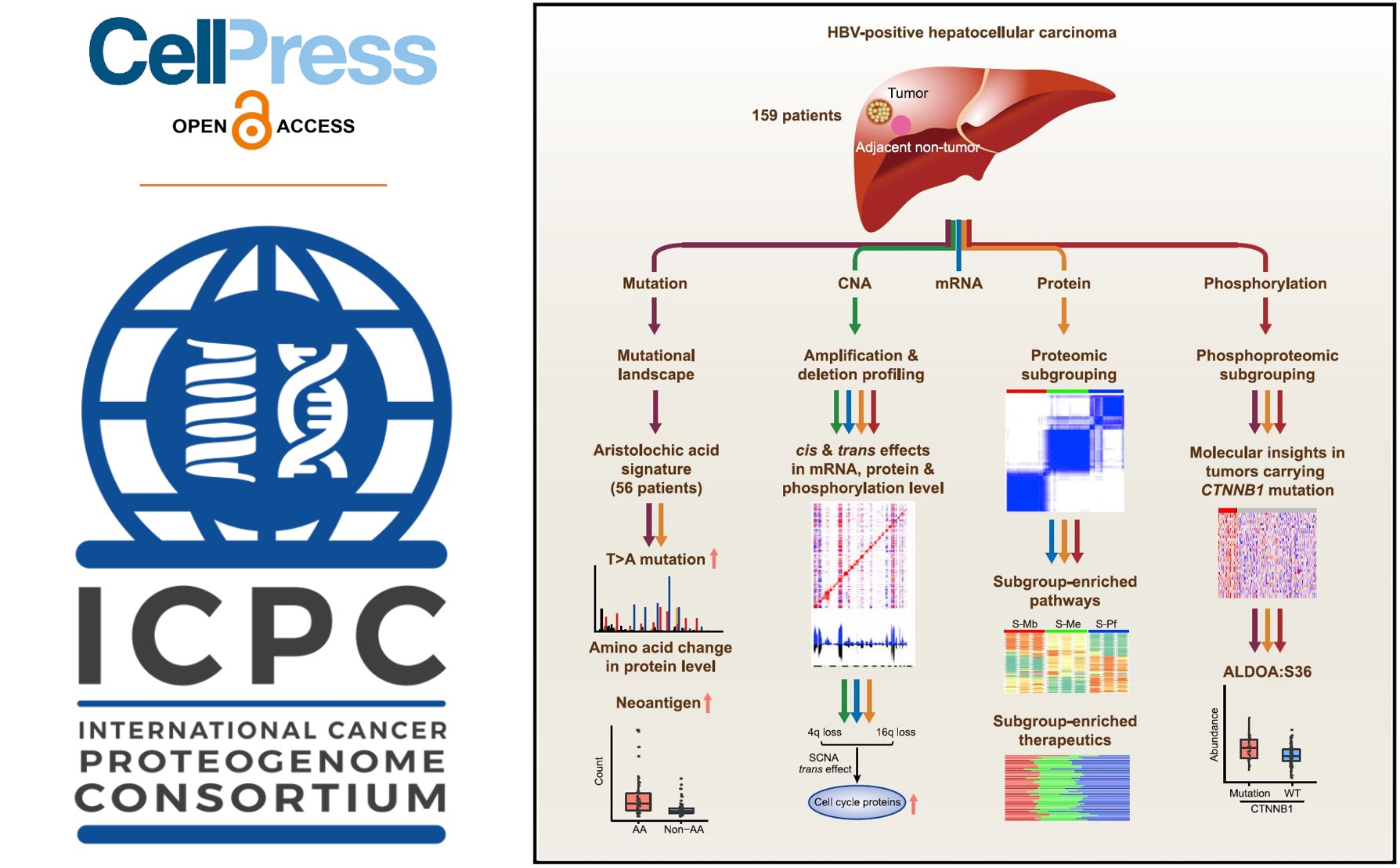 form of liver cancer, occurs most often in people with hepatitis B or C viral (HBV or HCV) infections who drink large amounts of alcohol. Unlike the recent progress of antiviral-eliminating therapy for HCV, current antiviral therapy is only able to reduce rather than eliminate HBV and is estimated to affect 292 million peop
form of liver cancer, occurs most often in people with hepatitis B or C viral (HBV or HCV) infections who drink large amounts of alcohol. Unlike the recent progress of antiviral-eliminating therapy for HCV, current antiviral therapy is only able to reduce rather than eliminate HBV and is estimated to affect 292 million peop


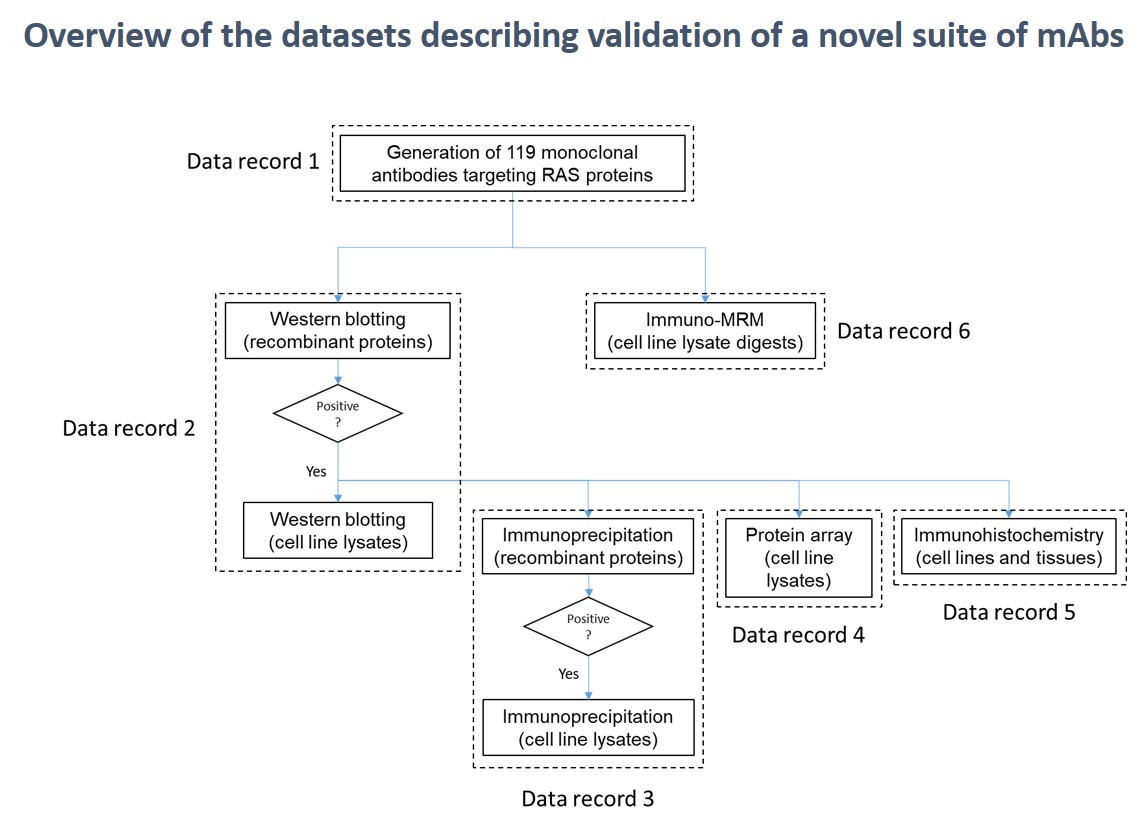
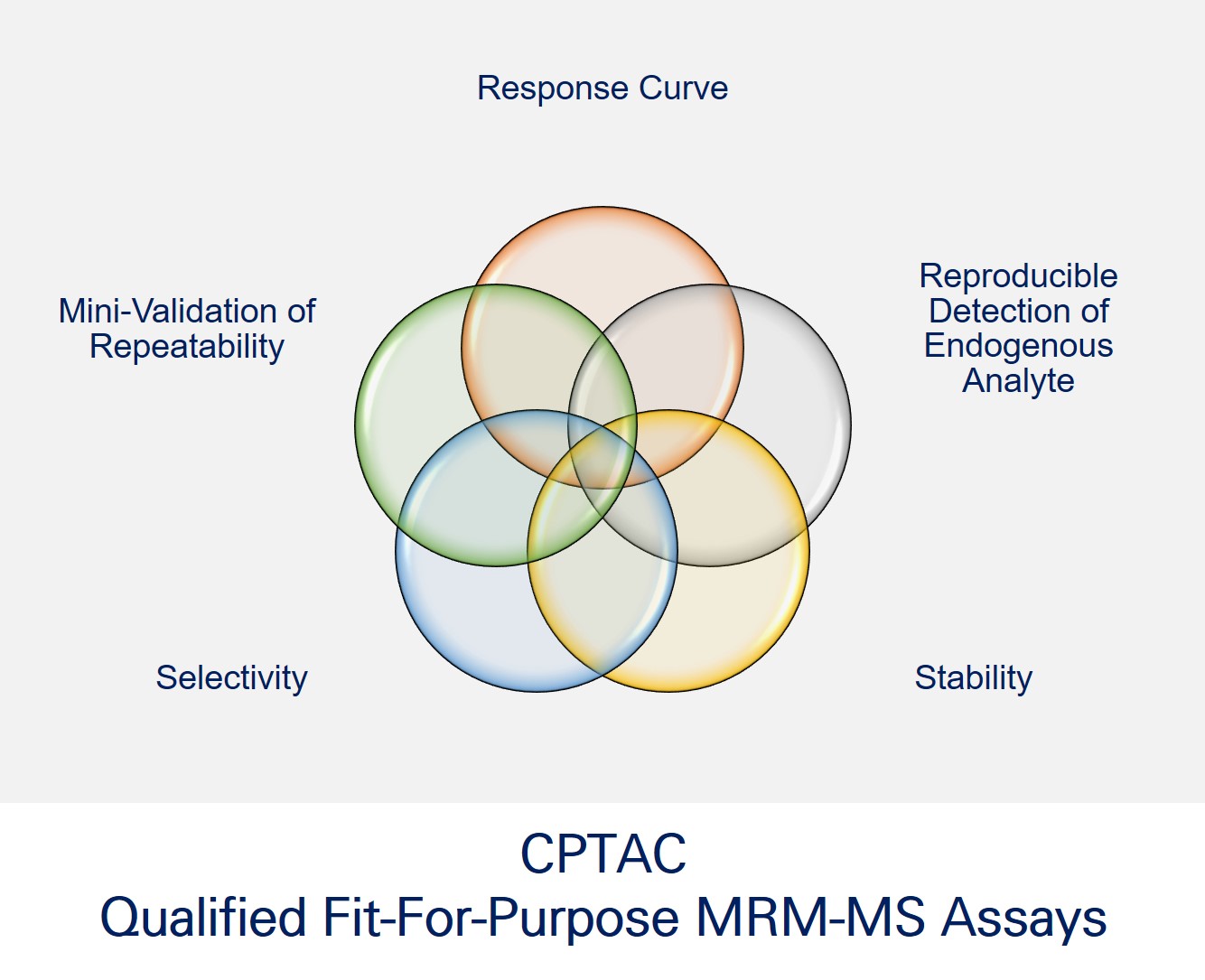 researchers from the University of Victoria – Genome British Columbia Proteomics Centre just released over 130 quantitative, fit-for-purpose, multiplexed mouse multiple reaction monitoring (MRM) mass spectrometry (MS)-based plasma assays for public use on NCI’s
researchers from the University of Victoria – Genome British Columbia Proteomics Centre just released over 130 quantitative, fit-for-purpose, multiplexed mouse multiple reaction monitoring (MRM) mass spectrometry (MS)-based plasma assays for public use on NCI’s 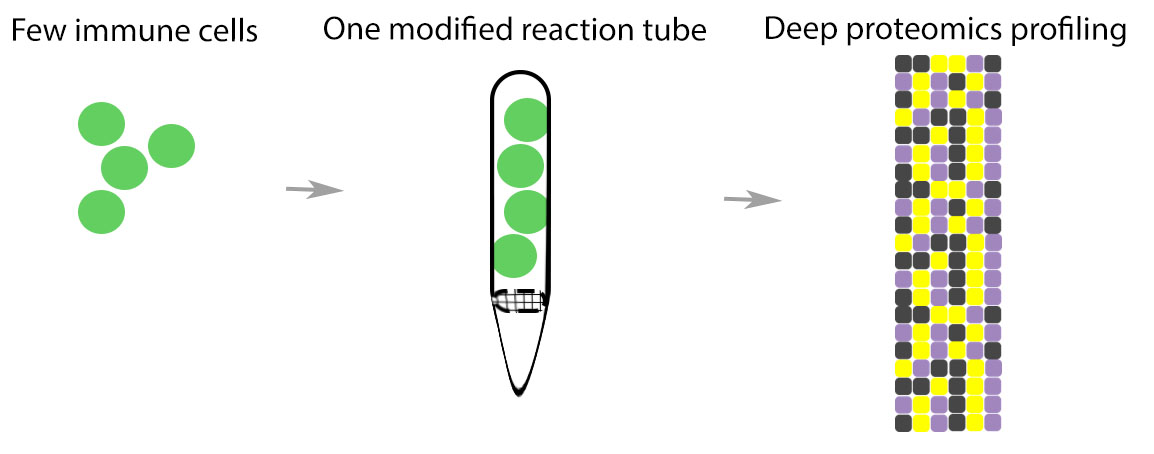 using a large sample input to explore the dynamic nature of protein expression and regulation. However, there are instances when researchers have the opposite due to prior methodology such as fluorescence activated cell sorting (FACS) or when dealing with patient samples.
using a large sample input to explore the dynamic nature of protein expression and regulation. However, there are instances when researchers have the opposite due to prior methodology such as fluorescence activated cell sorting (FACS) or when dealing with patient samples. the
the 